Histone: Difference between revisions
imported>Subpagination Bot m (Add {{subpages}} and remove any categories (details)) |
mNo edit summary |
||
| (4 intermediate revisions by one other user not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
'''Histone''' [[protein]]s are essential for the packaging of [[DNA]] into [[chromosome]]s within the [[nucleus]] of a [[cell]]. They also control, in some degree, the expression of proteins from DNA. Histone proteins play an important role in DNA packaging, [[chromosome]] stabilization and gene expression. Previously, it was thought that only five types of histones existed, namely H1, H2A, H2B, H3 and H4. The discovery of any new class of histones was virtually discarded by many, owing to the highly conserved nature of these proteins. However, recent research has revealed several new types of histone proteins which also play key roles in [[eukaryotic cells]]. DNA present in the nucleus of a cell is arranged in a compact and super-coiled manner to form chromosomes. This is called ‘DNA Packaging’. If this compactness is omitted, a human cell, for example, will find it awfully tricky to tackle a 1.5 cm long molecule of DNA. This DNA wrapping assists in reducing the dimensions of DNA from centimetre to micrometer in a cell. This 10,000 fold compression is achieved by virtue of the most conserved families of cellular proteins called Histones [1]. Even at this firmness, histone characteristics are such that chromosomes continue to maintain their dynamism. In following sections, topics pertaining to basic functions of histone and their recently discovered diversity have been discussed. | |||
Histone [[ | |||
DNA present in | |||
== Histones and their Functions == | == Histones and their Functions == | ||
[[Image:Chromatin Packaging.gif|right|thumb]] | [[Image:Chromatin Packaging.gif|right|thumb]] | ||
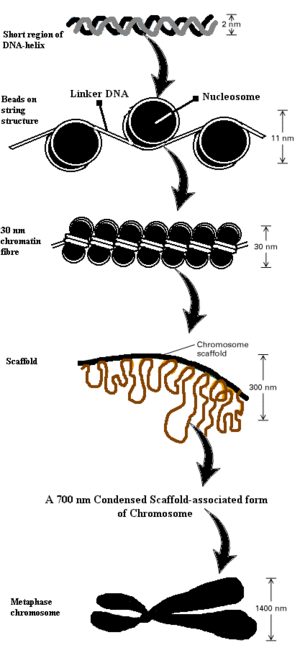
Eukaryotic DNA is bound tightly to an almost equal mass of | Eukaryotic DNA is bound tightly to an almost equal mass of histones, so that 300 million histone molecules may be found per human cell. Histones aid in fitting entire DNA content of a cell into its nucleus. Histones are principally of five types i.e. H1, H2A, H2B, H3 and H4. Out of these histones, H2A, H2B, H3 and H4 are involved in formation of the fundamental sub-unit of [[chromatin]] called a [[nucleosome]] (Figure 1). Nucleosome core consists of an 8 histone complex containing two molecules each of H2A, H2B, H3 and H4. When chromatin is extracted from cells at low salt concentrations, it resembles “beads on string”. In this outward appearance, the string is supposed to be DNA which connects bead-like nucleosomes. These nucleosomes have diameter of 10 nm. This octameric histone core(nucleosome) is roughly disk-shaped. Approximately 165 base pairs (bp) of DNA wrap about this nucleosome, taking about 2.85 turns around it [2]. A region called linker DNA separates each nucleosome from other succeeding or preceding nucleosomes. This linker DNA is stabilized by the fifth histone, H1. H1 binds to each nucleosome and its adjacent linker DNA to assist in bringing adjoining nucleosomes together, for further super coiling. This level of compactness gives rise to a 30 nm diameter chromatin fibre. In our cells, newly replicated DNA is integrated with respective nucleosomes as soon as the DNA replication fork has passed a particular position. Many proteins, which help in assembling DNA with nucleosomes, have been characterized ''in vitro'' and ''in vivo''. The 30 nm fibre thus formed then condenses into a spiral or solenoid arrangement with six nucleosomes per turn. This fibre further condenses to form a chromosome of diameter 1,400 nm. An array of acetylation, methylation and phosphorylation on particular [[amino acid]] residues in the histones plays an extremely vital role in the regulation and expression of genes [3]. | ||
== Histones in Chromosomes == | == Histones in Chromosomes == | ||
Chromosomal histones | Chromosomal histones are one of the most highly conserved protein molecules in eukaryotic cells. Such is their conservation that, amino acid sequences of histone H3 from garden [[plant]] and calf [[thymus]] differs only at 2 of 102 positions; similarly sequences of H3 protein differ by only one amino-acid residue between sea-urchin tissue and calf thymus. This implies a amalgamated role of all amino acids present in these histones. Pioneering glimpses of these basic histones in eukaryotic cells came in 1997 when [[X-ray diffraction]] revealed their organisation [4]. Histone proteins are rich in positively-charged basic amino acids. These basic amino acids interact with DNA, which is negatively-charged. All such histones have been found to be small, 102-135 amino acid-long molecules sharing a structural [[motif]] called the [[histone fold]], consisting of three [[α-helices]]. Other than histone folds, each core histone protein possesses a long N-terminal “tail”, extending out from the DNA-histone core. Post-translational modifications in histone proteins may cause conversion of some positively charged amino acids into negatively charged ones. Out of all of the histones associated with eukaryotic chromosomes, the H1 amino acid sequences vary the most from organism to organism. The high level of conservation among histone protein sequences was responsible for the hypothesis that variants of these proteins cannot exist, since ''in vitro'' experiments on [[yeast]]s had demonstrated that any change in histone sequences was lethal for the cell [1]. However, recent research has refuted this idea; numerous histone variants have been discovered recently which play vital roles in proper cell functioning. | ||
== The Modified Histones == | == The Modified Histones == | ||
Modified histones or histone-variants have been found to play crucial role in organisation of [[centromeres]], [[telomeres]] and inactive [[X-chromosomes]]. Inactive X-chromosome is also known as [[Barr body]]. Barr bodies are condensed mass of chromatin found in the nuclei of placental [[mammals]] containing one or more X- | Modified histones or histone-variants have been found to play a crucial role in the organisation of [[centromeres]], [[telomeres]] and inactive [[X-chromosomes]]. Inactive X-chromosome is also known as [[Barr body]]. Barr bodies are a condensed mass of chromatin found in the nuclei of placental [[mammals]] containing one or more X-chromosomes for dosage compensation [1]. Mammalian inactive X-chromosomes have been found to contain high levels of the histone-variant called [[macroH2A]] [5]. MacroH2A is nearly 3 times larger than normal H2A. The [[sea urchin]], on the other hand, has five versions of H2A variants, each of which gets expressed during different times in sea urchin development. Centromeres are specific sites on chromosomes where, spindle-fibre attachment occurs during cell division. Centromeres of vertebrate chromosomes and Drosophila chromosomes have been found to contain a variant of H3 histone, called [[CENP-A]]. Researchers have proposed this modification to be a very ancient evolutionary adaptation. CENP-A plays an important role in the cell cycle by associating with other proteins like [[CENP-B]], [[CENP-C]] and an array of miscellaneous molecules, to enable orderly separation of two [[chromatid]]s of chromosomes during cell-division [6]. Recent research has also been successful in pointing out histone-variants in highly conserved structure of nucleosomes. H2A and H3 histones, which were considered to be highly preserved, have also been shown to exist in variable forms at several instances. H2A has been shown to exist as H2AZ and H2AX, while H3 has been found to exist as H3.3 histone. Functionally, H2AZ replaces H2A, non-randomly and associates with reduced nucleosome stability. H2AX on the other hand, is relatively a more abundant histone-variant, as it has been found to play vital role in DNA breakage repair and V(D)J recombination event, which helps in generating immunoglobulin diversity [6]. Recently, H2AX protein has been found to play an important role in cell-cycle checkpoint mediation, which may be related to cancer induced due to DNA damage [7]. Histone H3.3 may be found incorporated in chromosomes of non-dividing cells and assist in functioning of transcriptionally active genes [8]. All these diverse histone-variants play crucial role in fate specific genetic expression and DNA stabilization in eukaryotic cells. | ||
== A Novel Histone - H5 == | |||
Discovery of histone variants arrived as a big surprise for many researchers, but an even more unanticipated discovery was that of an entirely new class of histone protein. Extensive research into histone protein evolution and diversity revealed that in certain tissues, H1 was replaced by special histones. These histones were called H5 histones and were discovered for the first time in chicken [[erythrocyte]]s [9]. Later, H5 histones were discovered in many nucleated red blood cells of birds, pointing to the fact that H5 may be involved in extensive silencing of chromosomal DNA in condensed nuclei of avian nucleated RBCs. Research has also elucidated that the structure of histone H5 exhibits similarity to the DNA-binding domain of the catabolite gene activator (CAP) protein [10]. Further research on evolution of H5 protein is underway. | |||
| Line 53: | Line 43: | ||
[9] Aviles F J, Chapman G E et al. (1978) The conformation of histone H5. Isolation and characterisation of the globular segment, Europ. J. Biochem., 88, 363-371. | [9] Aviles F J, Chapman G E et al. (1978) The conformation of histone H5. Isolation and characterisation of the globular segment, Europ. J. Biochem., 88, 363-371. | ||
[10] Ramakrishnan V, Finch J T et al. (2001) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219-223. | [10] Ramakrishnan V, Finch J T et al. (2001) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219-223.[[Category:Suggestion Bot Tag]] | ||
Latest revision as of 06:00, 28 August 2024
Histone proteins are essential for the packaging of DNA into chromosomes within the nucleus of a cell. They also control, in some degree, the expression of proteins from DNA. Histone proteins play an important role in DNA packaging, chromosome stabilization and gene expression. Previously, it was thought that only five types of histones existed, namely H1, H2A, H2B, H3 and H4. The discovery of any new class of histones was virtually discarded by many, owing to the highly conserved nature of these proteins. However, recent research has revealed several new types of histone proteins which also play key roles in eukaryotic cells. DNA present in the nucleus of a cell is arranged in a compact and super-coiled manner to form chromosomes. This is called ‘DNA Packaging’. If this compactness is omitted, a human cell, for example, will find it awfully tricky to tackle a 1.5 cm long molecule of DNA. This DNA wrapping assists in reducing the dimensions of DNA from centimetre to micrometer in a cell. This 10,000 fold compression is achieved by virtue of the most conserved families of cellular proteins called Histones [1]. Even at this firmness, histone characteristics are such that chromosomes continue to maintain their dynamism. In following sections, topics pertaining to basic functions of histone and their recently discovered diversity have been discussed.
Histones and their Functions
Eukaryotic DNA is bound tightly to an almost equal mass of histones, so that 300 million histone molecules may be found per human cell. Histones aid in fitting entire DNA content of a cell into its nucleus. Histones are principally of five types i.e. H1, H2A, H2B, H3 and H4. Out of these histones, H2A, H2B, H3 and H4 are involved in formation of the fundamental sub-unit of chromatin called a nucleosome (Figure 1). Nucleosome core consists of an 8 histone complex containing two molecules each of H2A, H2B, H3 and H4. When chromatin is extracted from cells at low salt concentrations, it resembles “beads on string”. In this outward appearance, the string is supposed to be DNA which connects bead-like nucleosomes. These nucleosomes have diameter of 10 nm. This octameric histone core(nucleosome) is roughly disk-shaped. Approximately 165 base pairs (bp) of DNA wrap about this nucleosome, taking about 2.85 turns around it [2]. A region called linker DNA separates each nucleosome from other succeeding or preceding nucleosomes. This linker DNA is stabilized by the fifth histone, H1. H1 binds to each nucleosome and its adjacent linker DNA to assist in bringing adjoining nucleosomes together, for further super coiling. This level of compactness gives rise to a 30 nm diameter chromatin fibre. In our cells, newly replicated DNA is integrated with respective nucleosomes as soon as the DNA replication fork has passed a particular position. Many proteins, which help in assembling DNA with nucleosomes, have been characterized in vitro and in vivo. The 30 nm fibre thus formed then condenses into a spiral or solenoid arrangement with six nucleosomes per turn. This fibre further condenses to form a chromosome of diameter 1,400 nm. An array of acetylation, methylation and phosphorylation on particular amino acid residues in the histones plays an extremely vital role in the regulation and expression of genes [3].
Histones in Chromosomes
Chromosomal histones are one of the most highly conserved protein molecules in eukaryotic cells. Such is their conservation that, amino acid sequences of histone H3 from garden plant and calf thymus differs only at 2 of 102 positions; similarly sequences of H3 protein differ by only one amino-acid residue between sea-urchin tissue and calf thymus. This implies a amalgamated role of all amino acids present in these histones. Pioneering glimpses of these basic histones in eukaryotic cells came in 1997 when X-ray diffraction revealed their organisation [4]. Histone proteins are rich in positively-charged basic amino acids. These basic amino acids interact with DNA, which is negatively-charged. All such histones have been found to be small, 102-135 amino acid-long molecules sharing a structural motif called the histone fold, consisting of three α-helices. Other than histone folds, each core histone protein possesses a long N-terminal “tail”, extending out from the DNA-histone core. Post-translational modifications in histone proteins may cause conversion of some positively charged amino acids into negatively charged ones. Out of all of the histones associated with eukaryotic chromosomes, the H1 amino acid sequences vary the most from organism to organism. The high level of conservation among histone protein sequences was responsible for the hypothesis that variants of these proteins cannot exist, since in vitro experiments on yeasts had demonstrated that any change in histone sequences was lethal for the cell [1]. However, recent research has refuted this idea; numerous histone variants have been discovered recently which play vital roles in proper cell functioning.
The Modified Histones
Modified histones or histone-variants have been found to play a crucial role in the organisation of centromeres, telomeres and inactive X-chromosomes. Inactive X-chromosome is also known as Barr body. Barr bodies are a condensed mass of chromatin found in the nuclei of placental mammals containing one or more X-chromosomes for dosage compensation [1]. Mammalian inactive X-chromosomes have been found to contain high levels of the histone-variant called macroH2A [5]. MacroH2A is nearly 3 times larger than normal H2A. The sea urchin, on the other hand, has five versions of H2A variants, each of which gets expressed during different times in sea urchin development. Centromeres are specific sites on chromosomes where, spindle-fibre attachment occurs during cell division. Centromeres of vertebrate chromosomes and Drosophila chromosomes have been found to contain a variant of H3 histone, called CENP-A. Researchers have proposed this modification to be a very ancient evolutionary adaptation. CENP-A plays an important role in the cell cycle by associating with other proteins like CENP-B, CENP-C and an array of miscellaneous molecules, to enable orderly separation of two chromatids of chromosomes during cell-division [6]. Recent research has also been successful in pointing out histone-variants in highly conserved structure of nucleosomes. H2A and H3 histones, which were considered to be highly preserved, have also been shown to exist in variable forms at several instances. H2A has been shown to exist as H2AZ and H2AX, while H3 has been found to exist as H3.3 histone. Functionally, H2AZ replaces H2A, non-randomly and associates with reduced nucleosome stability. H2AX on the other hand, is relatively a more abundant histone-variant, as it has been found to play vital role in DNA breakage repair and V(D)J recombination event, which helps in generating immunoglobulin diversity [6]. Recently, H2AX protein has been found to play an important role in cell-cycle checkpoint mediation, which may be related to cancer induced due to DNA damage [7]. Histone H3.3 may be found incorporated in chromosomes of non-dividing cells and assist in functioning of transcriptionally active genes [8]. All these diverse histone-variants play crucial role in fate specific genetic expression and DNA stabilization in eukaryotic cells.
A Novel Histone - H5
Discovery of histone variants arrived as a big surprise for many researchers, but an even more unanticipated discovery was that of an entirely new class of histone protein. Extensive research into histone protein evolution and diversity revealed that in certain tissues, H1 was replaced by special histones. These histones were called H5 histones and were discovered for the first time in chicken erythrocytes [9]. Later, H5 histones were discovered in many nucleated red blood cells of birds, pointing to the fact that H5 may be involved in extensive silencing of chromosomal DNA in condensed nuclei of avian nucleated RBCs. Research has also elucidated that the structure of histone H5 exhibits similarity to the DNA-binding domain of the catabolite gene activator (CAP) protein [10]. Further research on evolution of H5 protein is underway.
References
[1] Alberts B et al. (2001) Molecular Biology of the Cell-4th Ed., Garland Science Publication, New York.
[2] Kornberg R D (1974) Chromatin structure: A repeating unit of histones and DNA. Science, 184, 868-871.
[3] Goll M G & Bestor T H (2002) Histone modification and replacement in chromatin activation. Genes Dev., 16, 1739-1742.
[4] Lunger K, Mander A W et al. (1997) Crystal structure of the nucleosome core particle at 2.8 A0 resolution. Nature, 389, 251-260.
[5] Chadwick B P & Willard H F (2002) Cell cycle-dependant localization of macroH2A in chromatin of the inactive X Chromosome. J. Cell Biol., 157, 1113-1123.
[6] Redon C et al. (2002) Histone H2A variants H2AX and H2AZ. Curr. Opin. Genet. Dev., 12, 162-169.
[7] Kastan M B & Bartek J (2004) Cell-cycle checkpoints and cancer. Nature, 432, 316-323.
[8] Ahmad K & Henikoff S (2002) Histone H3 variants specify modes of chromatin assembly. Proc. Nat'l. Acad. Sci. USA, 10.1073/pnas.172403699, 2002.
[9] Aviles F J, Chapman G E et al. (1978) The conformation of histone H5. Isolation and characterisation of the globular segment, Europ. J. Biochem., 88, 363-371.
[10] Ramakrishnan V, Finch J T et al. (2001) Crystal structure of globular domain of histone H5 and its implications for nucleosome binding. Nature, 362, 219-223.