Protein structure: Difference between revisions
imported>David E. Volk (decent stub, images to follow) |
mNo edit summary |
||
| (28 intermediate revisions by 6 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | |||
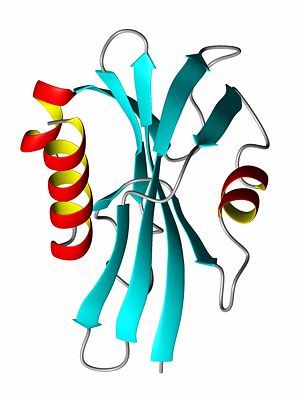
{{Image|ProteinRibbonByDEVolk.jpg|right|300px|Ribbon diagram of the P13 protein}} | |||
Proteins participate in numerous functions within living organisms, and protein function is largely dependent on the '''protein structure'''. Many diseases are caused by proteins whose structures have been modified, either due to amino acid substitutions (sicle cell anemia, cancer) resulting from DNA changes or by incorrect [[protein folding]] (alzheimer's). | Proteins participate in numerous functions within living organisms, and protein function is largely dependent on the '''protein structure'''. Many diseases are caused by proteins whose structures have been modified, either due to amino acid substitutions (sicle cell anemia, cancer) resulting from DNA changes or by incorrect [[protein folding]] (alzheimer's). | ||
== Basis of protein structure == | == Basis of protein structure == | ||
A Protein is a linear polymer of different [[amino acid|amino acids]], whose properties vary. To a large degree the shape of a protein is determined by the hydrophobic | A Protein is a linear polymer of different [[amino acid|amino acids]], whose properties vary. To a large degree the shape of a protein is determined by the [[hydrophobic]] amino acids which prefer not to be solvated and by the [[hydrophilic]] amino acids, which like to be exposed to water. Proteins typically fold up in such as way to minimize the solvent exposure of the hydrophobic amino acids while exposing the hydrophilic ones. This is accomplished by both local [[#secondary structure|secondary structure]] and by the overall tertiary structure of the protein. Hydrogen bonding also plays a significant role in a protein's structure, especially in regards to the [[secondary structure]] elements. Some proteins, often those with a very high percentage of hydrophilic amino acids, are natively unfolded, or are said to have a [[random coil]] structure. For example, many hormone receptor proteins, or large portions of them, are natively unfolded. Most of the proteins for which structures has been determined have secondary structures consisting largely of [[alpha helix|alpha helices]] and/or [[beta-sheet]]s which are tighly packed together to bury the hydrophobic amino acids (typically [[aliphatic]] and [[aromatic]] amino acids) away from the solvent. | ||
== | ==Primary structure== | ||
In a protein, primary structure is a term that refers to the linear sequence of amino acids that make up the protein. The structure of a protein is mostly dictated by its primary sequence. Related protein sequences can often have a high degree of [[sequence identity]], meaning that when the protein sequences are aligned, they have the same amino acid at many positions, and an even higher level of sequence homology, in which other positions have different, but very similar, amino acids. | |||
==Secondary structure== | |||
=== Alpha helix structure === | |||
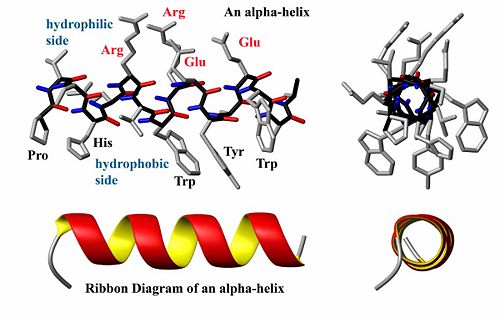
{{Image|AlphaHelixByDEVolk.jpg|right|500px|Structure and ribbon diagram of a protein alpha helix. The backbone atoms form a coil (black bonds) while the carbonyl groups (red) form hydrogen bonds with the amide groups (blue)}} | |||
A helix is generally a <sub>3</sub>10-helix (seldom) or an [[alpha-helix]], depending on the degree of twist in the protein backbone. By coiling the backbone atoms (black in the figure) to form a helix, a protein forms energetically favorable hydrogen bonds between the carboxyl oxygen atoms (end of red bonds) at a particular position (say <b>i</b> for example) with the amide proton atom (end of blue bonds) at position <b>i+4</b> (see figure). This formation also minimizes steric interactions between adjacent side chains. When two helices are packed against each other, they often point in opposite directions to reduce the dipole-dipole interaction between them. The dipole of each helix is caused by the alignment of the polar bonds within a helix. Often, one side of a helix is hydrophobic while the other side is hydrophilic, and such a helix is said to be amphiphilic. Some proteins are made entirely or nearly entirely of only alpha helices. Musle tissue and keratin are two examples. | |||
=== Beta sheet structure === | |||
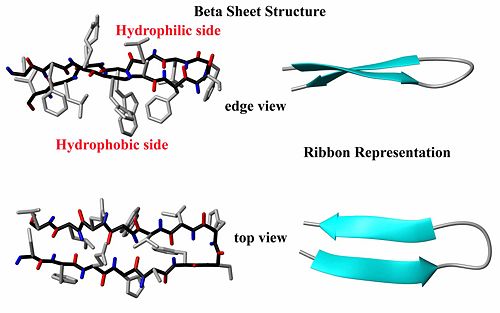
{{Image|BetaSheetByDEVolk.jpg|right|500px|Structure and ribbon diagram of a protein beta sheet. Note the oscillating positions of the carboxyl (red) and amide (blue) groups which form hydrogen bonds between the two beta strands.}} | |||
The other major element of secondary structure is the [[beta-sheet]] which is comprised of individual [[beta-strand]]s. In a beta-strand, the protein backbone atoms (N,Ca and C) form a zig-zag pattern (see figure). The result is that both the carboxyl group oxygen atoms and the amide group protons alternative back and forth on the strand. When two beta-strands interact, favorable hydrogen bonds can be formed between the carboxyl oxygen atoms and the amide group protons, thus stabilizing the structure. Like helices, beta-strands often have a hydrophilic side and a hydrophobic side, so that interactions between them and other secondary structure elements can be used to bury the hydrophobic amino acids and leave the hydrophilic amino acids exposed. The ends of beta-strands often end with beta-turns (read below). There are four types of beta-turns, called Types I, II, I' and II'. I' and II' are mirror images of types I and II. The ends of beta-strand may be flexible loop structures as well. | |||
== Tertiary structure == | == Tertiary structure == | ||
A protein's [[tertiary structure]] describes how all of the secondary structure elements fold upon one another. In order to bury the hydrophobic amino acid side chains (or residual groups), the secondary structural elements collapse upon each other so that most of the hydrophobic side chains are secluded away from the solvent, while the hydrophlic amino acids are exposed to the solvent. Thus, a beta-sheet might be protected on both sides by alpha-helices such that the polar side of the helix is exposed to the solvent while the non-polar, hydrophic side of the helix interacts with the hydrophobic residues of the beta-sheet. | A protein's [[tertiary structure]] (see first image) describes how all of the secondary structure elements fold upon one another. In order to bury the hydrophobic amino acid side chains (or residual groups), the secondary structural elements collapse upon each other so that most of the hydrophobic side chains are secluded away from the solvent, while the hydrophlic amino acids are exposed to the solvent. Thus, a beta-sheet might be protected on both sides by alpha-helices such that the polar side of the helix is exposed to the solvent while the non-polar, hydrophic side of the helix interacts with the hydrophobic residues of the beta-sheet. | ||
== Quaternary structure == | == Quaternary structure == | ||
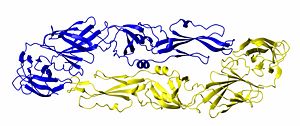
{{Image|DimerByDEVolk.jpg|right|300px|Structure of the Dengue virus envelope protein dimer. One monomer is shown in blue and the other is shown in gold.}} | |||
Quaternary structure describes the interaction (structure) of a group of proteins. Many proteins exist in dimer form, meaning that two molecules of protein bind to each other. Many DNA binding proteins are homodimers. Other proteins form homotrimers and homotetramers. If different protein bind together, they are referred to as hetero-dimers, hetero-trimers, and so forth. Many biological processes occur when dozens of proteins or proteins/DNA/RNA molecules bind together to form a quaternary structure. | |||
===Domains=== | |||
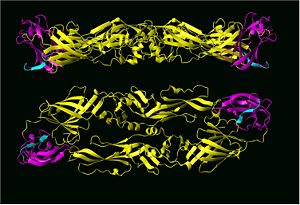
{{Image|Dengue domain III CZ.jpg|right|300px|Top and side view of the Dengue 2 virus envelope protein dimer structure with domain III hi-lighted in magenta and cyan.}} | |||
Many proteins, especially large ones, contain distinct, stable structural domains that are interconnected by linker peptides. The dengue-2 virus envelope protein, for example, contatins three distinct structural domains, referred to as domains I, II and III, that each play a different role in virus infectivity. Domains within a single protein often rearrange their relative positions with each other when a protein binds to its native ligand, cells or other proteins. When dengue viruses bind to cells for example, not only is does the tertiary structure change, that is protein dimers change to protein trimers, the relative orientation of domains I and III are greatly changed within each single protein. | |||
== Protein superstructures== | |||
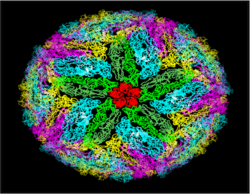
{{Image|Flaviball1-CZ.png|right|250px|Protein dimers form the flavivirus particle outer surface. View looking at a five-fold symmetric axis.}} | |||
Some proteins form superstructures. Typical examples of this would be cell membranes and viral structural proteins. In enveloped viruses, for example, hundreds of proteins form a outer barrier through dimer-dimer contacts. Biological cell mambranes are formed by bilayers in which amphiphilic proteins, containing hydrophobic and hydrophilic ends, form large layers such that their hydrophobic ends form the center of the bilayer while the hydrohilic head groups form the outer and inner edges facing aqueous environments. | |||
===Motifs=== | |||
==Misfolding and disease== | |||
===Chaperones=== | |||
In the course of biological processes, some proteins become misfolded. A special set of proteins called [[chaperone (protein)|chaperones]] are specialized proteins that refold other misfolded proteins. The GroEL/GroES system is perhaps the best studied of these special proteins. Typically, chaperones form a large crockpot-like container by the interaction of seven or more individual proteins, as well as a "lid" also formed from multiple proteins. The misfolded protein enteres the open chaperone structure, which is then capped by the lid. The entire process is mediated by an ATP/ADP-controlled process. Basically, the inside of the chaperone container pulls the misfolded protein apart and then allows it to refold itself. | |||
===Prions=== | |||
===Sickle cell anemia=== | |||
== Methods used to determine protein structure == | |||
* [[X-ray crystallography]] | |||
* [http://en.citizendium.org/wiki/NMR_spectroscopy NMR spectroscopy] | |||
* Circular Dichroism (CD) | |||
* Infrared (IR)spectroscopy | |||
== References == | == References == | ||
<references/>[[Category:Suggestion Bot Tag]] | |||
[[Category: | |||
Latest revision as of 16:01, 7 October 2024
Proteins participate in numerous functions within living organisms, and protein function is largely dependent on the protein structure. Many diseases are caused by proteins whose structures have been modified, either due to amino acid substitutions (sicle cell anemia, cancer) resulting from DNA changes or by incorrect protein folding (alzheimer's).
Basis of protein structure
A Protein is a linear polymer of different amino acids, whose properties vary. To a large degree the shape of a protein is determined by the hydrophobic amino acids which prefer not to be solvated and by the hydrophilic amino acids, which like to be exposed to water. Proteins typically fold up in such as way to minimize the solvent exposure of the hydrophobic amino acids while exposing the hydrophilic ones. This is accomplished by both local secondary structure and by the overall tertiary structure of the protein. Hydrogen bonding also plays a significant role in a protein's structure, especially in regards to the secondary structure elements. Some proteins, often those with a very high percentage of hydrophilic amino acids, are natively unfolded, or are said to have a random coil structure. For example, many hormone receptor proteins, or large portions of them, are natively unfolded. Most of the proteins for which structures has been determined have secondary structures consisting largely of alpha helices and/or beta-sheets which are tighly packed together to bury the hydrophobic amino acids (typically aliphatic and aromatic amino acids) away from the solvent.
Primary structure
In a protein, primary structure is a term that refers to the linear sequence of amino acids that make up the protein. The structure of a protein is mostly dictated by its primary sequence. Related protein sequences can often have a high degree of sequence identity, meaning that when the protein sequences are aligned, they have the same amino acid at many positions, and an even higher level of sequence homology, in which other positions have different, but very similar, amino acids.
Secondary structure
Alpha helix structure
A helix is generally a 310-helix (seldom) or an alpha-helix, depending on the degree of twist in the protein backbone. By coiling the backbone atoms (black in the figure) to form a helix, a protein forms energetically favorable hydrogen bonds between the carboxyl oxygen atoms (end of red bonds) at a particular position (say i for example) with the amide proton atom (end of blue bonds) at position i+4 (see figure). This formation also minimizes steric interactions between adjacent side chains. When two helices are packed against each other, they often point in opposite directions to reduce the dipole-dipole interaction between them. The dipole of each helix is caused by the alignment of the polar bonds within a helix. Often, one side of a helix is hydrophobic while the other side is hydrophilic, and such a helix is said to be amphiphilic. Some proteins are made entirely or nearly entirely of only alpha helices. Musle tissue and keratin are two examples.
Beta sheet structure
The other major element of secondary structure is the beta-sheet which is comprised of individual beta-strands. In a beta-strand, the protein backbone atoms (N,Ca and C) form a zig-zag pattern (see figure). The result is that both the carboxyl group oxygen atoms and the amide group protons alternative back and forth on the strand. When two beta-strands interact, favorable hydrogen bonds can be formed between the carboxyl oxygen atoms and the amide group protons, thus stabilizing the structure. Like helices, beta-strands often have a hydrophilic side and a hydrophobic side, so that interactions between them and other secondary structure elements can be used to bury the hydrophobic amino acids and leave the hydrophilic amino acids exposed. The ends of beta-strands often end with beta-turns (read below). There are four types of beta-turns, called Types I, II, I' and II'. I' and II' are mirror images of types I and II. The ends of beta-strand may be flexible loop structures as well.
Tertiary structure
A protein's tertiary structure (see first image) describes how all of the secondary structure elements fold upon one another. In order to bury the hydrophobic amino acid side chains (or residual groups), the secondary structural elements collapse upon each other so that most of the hydrophobic side chains are secluded away from the solvent, while the hydrophlic amino acids are exposed to the solvent. Thus, a beta-sheet might be protected on both sides by alpha-helices such that the polar side of the helix is exposed to the solvent while the non-polar, hydrophic side of the helix interacts with the hydrophobic residues of the beta-sheet.
Quaternary structure
Quaternary structure describes the interaction (structure) of a group of proteins. Many proteins exist in dimer form, meaning that two molecules of protein bind to each other. Many DNA binding proteins are homodimers. Other proteins form homotrimers and homotetramers. If different protein bind together, they are referred to as hetero-dimers, hetero-trimers, and so forth. Many biological processes occur when dozens of proteins or proteins/DNA/RNA molecules bind together to form a quaternary structure.
Domains
Many proteins, especially large ones, contain distinct, stable structural domains that are interconnected by linker peptides. The dengue-2 virus envelope protein, for example, contatins three distinct structural domains, referred to as domains I, II and III, that each play a different role in virus infectivity. Domains within a single protein often rearrange their relative positions with each other when a protein binds to its native ligand, cells or other proteins. When dengue viruses bind to cells for example, not only is does the tertiary structure change, that is protein dimers change to protein trimers, the relative orientation of domains I and III are greatly changed within each single protein.
Protein superstructures
Some proteins form superstructures. Typical examples of this would be cell membranes and viral structural proteins. In enveloped viruses, for example, hundreds of proteins form a outer barrier through dimer-dimer contacts. Biological cell mambranes are formed by bilayers in which amphiphilic proteins, containing hydrophobic and hydrophilic ends, form large layers such that their hydrophobic ends form the center of the bilayer while the hydrohilic head groups form the outer and inner edges facing aqueous environments.
Motifs
Misfolding and disease
Chaperones
In the course of biological processes, some proteins become misfolded. A special set of proteins called chaperones are specialized proteins that refold other misfolded proteins. The GroEL/GroES system is perhaps the best studied of these special proteins. Typically, chaperones form a large crockpot-like container by the interaction of seven or more individual proteins, as well as a "lid" also formed from multiple proteins. The misfolded protein enteres the open chaperone structure, which is then capped by the lid. The entire process is mediated by an ATP/ADP-controlled process. Basically, the inside of the chaperone container pulls the misfolded protein apart and then allows it to refold itself.
Prions
Sickle cell anemia
Methods used to determine protein structure
- X-ray crystallography
- NMR spectroscopy
- Circular Dichroism (CD)
- Infrared (IR)spectroscopy