Bifidobacterium bifidum: Difference between revisions
imported>Kelly Smith |
mNo edit summary |
||
| (38 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
{{Taxobox | {{Taxobox | ||
| Line 15: | Line 14: | ||
| binomial_authority = | | binomial_authority = | ||
}} | }} | ||
==Description and significance== | ==Description and significance== | ||
''Bifidobacterium bifidum'' is included in a subsection of [[bacteria]] known as [[probiotics]]. | ''Bifidobacterium bifidum'' is included in a subsection of [[bacteria]] known as [[probiotics]].<ref>[http://nccam.nih.gov/health/probiotics]</ref> These beneficial bacteria occur naturally within the human body and are found most commonly in the gastrointestinal tract.<ref>Willey, J.M., Sherwood, L.M., Woolverton, C.J. Prescott, Harley, and Klein's Microbiology. Seventh edition. 2008</ref> Bifidobacteria have also been detected, although significantly less in quantity, in breast milk, the mouth, and the vagina. While it is accepted that probiotics act opportunistically within the intestine, in effect beating out known pathogenic species, researchers continue to investigate the specific mechanisms by which bifidobacteria act. As a part of the probiotic group of bacteria, bifidobacteria are attributed with a vast array of beneficial physiologic effects. Some of the benefits associated with bifidobacteria include: improved digestion, augmentation of the immune system, efficient production of lactic and [[acetic acid]] without producing carbon dioxide, lower serum [[cholesterol]] levels, lower incidence of allergies, enhanced [[calcium]] absorption, better synthesis of B-complex vitamins, and further promotes anti-tumorgenic effects in cases of colon cancer.<ref>Chen, X., Jiang, H., Yang, Y., Liu, N. “[Effect of exopolysaccharide from Bifidobacterium bifidum on cell of gastric cancer and human telomerase reverse transcriptase]”. Wei Sheng Wu Xue Bao. January, 2009. Vol. 49, Issue 1. p. 117-22</ref> In addition, bifidobacteria have commonly been utilized in home remedies for diarrhea, [[vaginitis]], yeast infections, as well as [[irritable bowel syndrome]]. | ||
==Genome structure== | ==Genome structure== | ||
''B. bifidum'' has a circular chromosome with a G-C content of approximately 55-67%. Bifidobacterial species' genomes range in size from 1.9 to 2.9 Mb. Their number of rRNA operons also varies between one and five. Although a number of bifidobacterial plasmids have been sequenced, all but one, NCFB 1454, from ''B. bifidum'', are not known to encode for a phenotypic trait. | ''B. bifidum'' has a circular chromosome with a G-C content of approximately 55-67%. Bifidobacterial species' genomes range in size from 1.9 to 2.9 Mb. Their number of rRNA operons also varies between one and five.<ref>[http://microbewiki.kenyon.edu/index.php/Bifidobacterium]</ref> Although a number of bifidobacterial plasmids have been sequenced, all but one, NCFB 1454, from ''B. bifidum'', are not known to encode for a phenotypic trait.<ref>[http://www.ncbi.nlm.nih.gov/nuccore/83416324?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum]</ref> It is believed that NCFB 1454 encodes bacteriocin ''bifidocin B.''<ref>Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G.F., Chater, K.F., van Sinderen, D. “Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum”. Micobiol Mol Biol Rev. September, 2007. Vol. 71. p. 495-548</ref> [[Bacteriocin]]s are antibiotic substances produced by one bacteria that inhibit the growth and metabolic activity of closely related species. | ||
The following links to The National Center for Biotechnology Information (NCBI) are only some of the genetic sequences available from a vast number of ''B. bifidum'' genes and plasmids. | The following links to The National Center for Biotechnology Information (NCBI) are only some of the genetic sequences available from a vast number of ''B. bifidum'' genes and plasmids. The gene for the 16S ribosomal [[RNA]] is particularly noteworthy as this is the most common identifying factor in the determination of the ''B. bifidum'' species. | ||
[http://www.ncbi.nlm.nih.gov/nuccore/188593525?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum''Bifidobacterium bifidum gene for 16S ribosomal RNA] | |||
[http://www.ncbi.nlm.nih.gov/nuccore/83416324?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum''Bifidobacterium bifidum'' pB80 plasmid] | [http://www.ncbi.nlm.nih.gov/nuccore/83416324?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum''Bifidobacterium bifidum'' pB80 plasmid] | ||
| Line 36: | Line 33: | ||
[http://www.ncbi.nlm.nih.gov/nuccore/167369737?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum''Bifidobacterium bifidum'' JCM 1254 lacto-N-biosidase (lnbB) gene] | [http://www.ncbi.nlm.nih.gov/nuccore/167369737?ordinalpos=1&itool=EntrezSystem2.PEntrez.Sequence.Sequence_ResultsPanel.Sequence_RVDocSum''Bifidobacterium bifidum'' JCM 1254 lacto-N-biosidase (lnbB) gene] | ||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
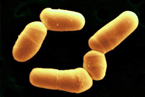
[[Image:NIHbifidum.jpg|150px|left]] | [[Image:NIHbifidum.jpg|150px|left]] | ||
Bifidobacteria are [[Gram-positive]], anaerobic, and | Bifidobacteria are [[Gram-positive]], anaerobic, nonmotile, and non spore forming. The characteristic rod or clubbed shape of ''B. bifidum'' varies from 0.5-1.3 μm x 1.5-8 μm. They can be found either living independently or associated in clusters and V-shaped pairs. These bacteria are commonly found curved and in a branched conformation. ''B. bifidum'' ferments oligosaccarides in the gastrointestinal tract. Approximately 10% of the total ''B. bifidum''genome is used to help the body properly breakdown and adsorb sugar.<ref>Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M.C., Desiere, F., Bork, P., Delley, M., Pridmore, R.D., Arigoni, F. "The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract". Proc Natl Acad Sci. 2002. Vol. 99. p. 14422-14427</ref> It does this by coding for ABC transporters, permeases, and proton symporters as opposed to phosphoenolpyruvate-phosphotransferase systems.<ref>Santacruz, A., Marcos, A., Warnberg, J., Marti, A., Martin-Matillas, M., Campoy, C., Moreno, L.A., Veiga, O., Redondo-Figuero, C., Garagorri, J.M., Azcona, C., Delgado, M., Garcia-Fuentes, M., Collado, M.C., Sanz, Y. “Interplay between weight loss and gut microbiota composition in overweight adolescents”. Obesity. April, 2009. doi:10.1038/oby.2009.112</ref> | ||
'' | |||
==Ecology== | ==Ecology== | ||
Bifidobacteria constitute approximately 90% of the microbiota found in the intestines of breast-fed infants. The specific breakdown | Bifidobacteria constitute approximately 90% of the microbiota found in the intestines of breast-fed infants. Infants delivered via caesarean section and who are formula fed as opposed to vaginally delivered and breast-fed have significantly lower gut populations of bifidobacteria.<ref>Gronlund, M.M., Gueimonde, M., Laitinen, K., Kociubinski, G., Gronroos, T., Salminen, S., Isolauri, E. “Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the bifidobacterium microbiota in infants at risk of allergic disease”. Clinical and Experimental Allergy. August, 2007. Vol. 37. p. 1764-1772</ref> The specific breakdown of bifidobacterial species varies but the major contributors include: ''B. brevi'', ''B. longum'', ''B. pseudocatenulatum'', ''B. adolescentis'', ''B. pseudolongum'', and ''B. bifidum''. The number of bifidobacteria in the fecal flora of adults, however, is much lower around 3-6%. Numerous studies have been conducted that illustrate a variance of gut microbiota both by subject as well as with subject age. ''B. bifidum'' is also found, although to a much lesser extent, in breast milk, the mouth, and the vagina. | ||
==Economic Importance== | ==Economic Importance== | ||
[[Image:Yogurt.jpg|200px|right]] | [[Image:Yogurt.jpg|200px|right]] | ||
The numerous beneficial attributes of bifidobacteria have led to | The numerous beneficial attributes of bifidobacteria have led to an increased awareness and the growing popularity of probiotics. Television commercials for Activia yogurt featuring the actress Jamie Lee Curtis have gained much notoriety. Probiotic supplements are relatively inexpensive and easily available to consumers both online and at local pharmacies without a prescription. The growing market for probiotics has sparked an influx of diverse product offerings, which include: yogurt, yogurt drinks, capsule supplements, cereal, snack bars, pediatric-specific supplements, baby formula, and toothpaste. | ||
==Current Research== | ==Current Research== | ||
| Line 58: | Line 49: | ||
====Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity- a randomized, double-blind, placebo-controlled study.==== | ====Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity- a randomized, double-blind, placebo-controlled study.==== | ||
The effects of [[prebiotics]] such as galactooligosaccharides (GOS) and long-chain fructooligosaccharides (lcFOS) that encourage the proliferation of bifidobacteria were examined on maternal gut microbiota during the third trimester of pregnancy and following delivery. This study aimed to determine whether maternal microbiota are passed to newborns via breast-feeding, and, as a secondary outcome, whether maternal gut microbiota affect the fetal immune response. | The effects of [[prebiotics]] such as galactooligosaccharides (GOS) and long-chain fructooligosaccharides (lcFOS) that encourage the proliferation of bifidobacteria were examined on maternal gut microbiota during the third trimester of pregnancy and following delivery. This study aimed to determine whether maternal microbiota are passed to newborns via breast-feeding, and, as a secondary outcome, whether maternal gut microbiota affect the fetal immune response.<ref>Mouni, F., Aissi, E., Hernandez, J., Gorocica, P., Bouquelet, S., Zenteno, E., Lascurain, R., Garfias, Y. “Effect of Bifidobacterium bifidum DSM 20082 cytoplasmic fraction on human immune cells”. Immunological Investigations. 2009. Vol. 38. p. 104-115</ref> It is known that early infant gut microbiota influence an individual’s susceptibility to developing allergies later in life, although the exact reason and mechanism are unknown. The quantity of bifidobacteria and lactobacilli from the excised population was determined by fluorescent in situ hybridization and quantitative polymerase chain reaction in maternal and infant stool samples. The second objective, to examine the fetal immune response, utilized samples of cord blood by using flow cytometry and cytokine multiplex-array analysis. The results of this study relay that although GOS/lcFOS supplementation during pregnancy has a bifidogenic effect on maternal intestine micro flora, it is not passed on to newborns. Furthermore, there was no correlation between increased maternal bifidobacteria and enhanced fetal immune response.<ref>[http://www.ajcn.org/cgi/content/abstract/86/5/1426 Shadid, R., Haarman, M., Knol, J., Theis, W., Beermann, C., Rjosk-Dendorfer, D., Schendel, D., Koletzko, B., Krauss-Etschmann, S. “Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity-a randomized, double-blind, placebo-controlled study”. Am J Clin Nutr. 2009. Vol. 86. p. 1426-37]</ref> | ||
====Exploring the diversity of the bifidobacterial population in the human intestinal tract.==== | ====Exploring the diversity of the bifidobacterial population in the human intestinal tract.==== | ||
While the beneficial aspects of bifidobacteria are widely accepted, the diversity and specific composition of the human intestine micro flora are less understood. Researchers conducted this study to identify specific bifidobacterial populations found in human intestines and fecal samples. Their protocol included plating human intestinal mucosal and fecal samples on selective media and further analyzing molecular data of selected rRNA gene sequences of individual colonies. Their findings clearly indicated that the majority, 704 of the 900 isolated colonies, were bifidobacteria. They further identified the six major species of bifidobacteria isolated from the intestine: ''B. longum'', ''B. pseudocatenulatum'', ''B. adolescentis'', ''B. pseudolongum'', ''B. breve'', and ''B. bifidum'', and two species found primarily in fecal samples, B. dentium and B. animalis subp. lactis. Their research indicated a correlation between age and the microbiota distribution of the intestine. A small selection of species were found exclusively in the adult human gut, while other species were found widely distributed. The study uncovered significant variance between individuals in the composition of fecal samples and intestinal mucosal samples as well as mild variance within the same subject (intrasubject variability) in different regions of the intestine. There were a small number of bifidobacteria that indicated the capacity to broadly colonize, which was based on the number that were able to be isolated from wide ecological distributions. | While the beneficial aspects of bifidobacteria are widely accepted, the diversity and specific composition of the human intestine micro flora are less understood. Researchers conducted this study to identify specific bifidobacterial populations found in human intestines and fecal samples. Their protocol included plating human intestinal mucosal and fecal samples on selective media and further analyzing molecular data of selected rRNA gene sequences of individual colonies. Their findings clearly indicated that the majority, 704 of the 900 isolated colonies, were bifidobacteria. They further identified the six major species of bifidobacteria isolated from the intestine: ''B. longum'', ''B. pseudocatenulatum'', ''B. adolescentis'', ''B. pseudolongum'', ''B. breve'', and ''B. bifidum'', and two species found primarily in fecal samples, B. dentium and B. animalis subp. lactis. Their research indicated a correlation between age and the microbiota distribution of the intestine. A small selection of species were found exclusively in the adult human gut, while other species were found widely distributed. The study uncovered significant variance between individuals in the composition of fecal samples and intestinal mucosal samples as well as mild variance within the same subject (intrasubject variability) in different regions of the intestine. There were a small number of bifidobacteria that indicated the capacity to broadly colonize, which was based on the number that were able to be isolated from wide ecological distributions.<ref>[http://www.ncbi.nlm.nih.gov/sites/entrez Turroni, F., Foroni, E., Pizzetti, P., Giubellini, V., Ribbera, A., Merusi, P., Cagnasso, P., Bizzarri, B., de'Angelis, G.L., Shanahan, F., van Sinderen, D., Ventura, M. “Exploring the diversity of the bifidobacterial population in the human intestinal tract”. Appl Environ Microbiol. March, 2009. Vol. 75, Issue 6. p. 1534-45]</ref> | ||
====Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR.==== | ====Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR.==== | ||
The aim of this study was to determine if breast milk contains bifidobacteria and further if it can be transferred from mother to infant via breast-feeding. This study examined 23 newborn infants that were exclusively breast-fed. Fructose-6-phosphate phosphoketolase assays were used to identify to the genus level and 16S rRNA gene sequencing to the species level. Bifidobacteria samples in breast milk were identified by PCR-denaturing gradient gel electrophoresis and approximate number determined by quantitative real-time PCR. Bifidobacteria were found in samples of both maternal milk and infant fecal matter. Eight of 23 mother child pairs showed a direct correlation between maternal milk microbial composition and infant feces microbial composition. Thus, this study concluded that breast milk serves as the source of bifidobacteria for the infant gut. | The aim of this study was to determine if breast milk contains bifidobacteria and further if it can be transferred from mother to infant via breast-feeding. This study examined 23 newborn infants that were exclusively breast-fed. Fructose-6-phosphate phosphoketolase assays were used to identify to the genus level and 16S rRNA gene sequencing to the species level. Bifidobacteria samples in breast milk were identified by PCR-denaturing gradient gel electrophoresis and approximate number determined by quantitative real-time PCR. Bifidobacteria were found in samples of both maternal milk and infant fecal matter. Eight of 23 mother child pairs showed a direct correlation between maternal milk microbial composition and infant feces microbial composition. Thus, this study concluded that breast milk serves as the source of bifidobacteria for the infant gut.<ref>[http://www.ncbi.nlm.nih.gov/pubmed/19088308?ordinalpos=1&itool=EntrezSystem2.PEntrez.Pubmed.Pubmed_ResultsPanel.Pubmed_DiscoveryPanel.Pubmed_Discovery_RA&linkpos=3&log$=relatedarticles&logdbfrom=pubmed Martin, R., Jimenez, E., Heilig, H., Fernandez, L., Marin, M.L., Zoetendal, E.G., Rodriguez, J.M. “Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR”. Appl Environ Microbiol. February, 2009. Vol. 75, Issue 4. p. 965-9]</ref> | ||
==References== | ==References== | ||
{{reflist|2}}[[Category:Suggestion Bot Tag]] | |||
[ | |||
Latest revision as of 11:00, 18 July 2024
| Scientific classification | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||
| Binomial name | ||||||||||||||
Description and significance
Bifidobacterium bifidum is included in a subsection of bacteria known as probiotics.[1] These beneficial bacteria occur naturally within the human body and are found most commonly in the gastrointestinal tract.[2] Bifidobacteria have also been detected, although significantly less in quantity, in breast milk, the mouth, and the vagina. While it is accepted that probiotics act opportunistically within the intestine, in effect beating out known pathogenic species, researchers continue to investigate the specific mechanisms by which bifidobacteria act. As a part of the probiotic group of bacteria, bifidobacteria are attributed with a vast array of beneficial physiologic effects. Some of the benefits associated with bifidobacteria include: improved digestion, augmentation of the immune system, efficient production of lactic and acetic acid without producing carbon dioxide, lower serum cholesterol levels, lower incidence of allergies, enhanced calcium absorption, better synthesis of B-complex vitamins, and further promotes anti-tumorgenic effects in cases of colon cancer.[3] In addition, bifidobacteria have commonly been utilized in home remedies for diarrhea, vaginitis, yeast infections, as well as irritable bowel syndrome.
Genome structure
B. bifidum has a circular chromosome with a G-C content of approximately 55-67%. Bifidobacterial species' genomes range in size from 1.9 to 2.9 Mb. Their number of rRNA operons also varies between one and five.[4] Although a number of bifidobacterial plasmids have been sequenced, all but one, NCFB 1454, from B. bifidum, are not known to encode for a phenotypic trait.[5] It is believed that NCFB 1454 encodes bacteriocin bifidocin B.[6] Bacteriocins are antibiotic substances produced by one bacteria that inhibit the growth and metabolic activity of closely related species.
The following links to The National Center for Biotechnology Information (NCBI) are only some of the genetic sequences available from a vast number of B. bifidum genes and plasmids. The gene for the 16S ribosomal RNA is particularly noteworthy as this is the most common identifying factor in the determination of the B. bifidum species.
Bifidobacterium bifidum gene for 16S ribosomal RNA
Bifidobacterium bifidum pB80 plasmid
Bifidobacterium bifidum alpha-galactosidase (melA) gene
Bifidobacterium bifidum pyrF, pyrK, pyrDb, pyrE genes
Bifidobacterium bifidum NCIMB 41171 beta-galactosidase Bbg1 gene
Bifidobacterium bifidum JCM 1254 lacto-N-biosidase (lnbB) gene
Cell structure and metabolism
Bifidobacteria are Gram-positive, anaerobic, nonmotile, and non spore forming. The characteristic rod or clubbed shape of B. bifidum varies from 0.5-1.3 μm x 1.5-8 μm. They can be found either living independently or associated in clusters and V-shaped pairs. These bacteria are commonly found curved and in a branched conformation. B. bifidum ferments oligosaccarides in the gastrointestinal tract. Approximately 10% of the total B. bifidumgenome is used to help the body properly breakdown and adsorb sugar.[7] It does this by coding for ABC transporters, permeases, and proton symporters as opposed to phosphoenolpyruvate-phosphotransferase systems.[8]
Ecology
Bifidobacteria constitute approximately 90% of the microbiota found in the intestines of breast-fed infants. Infants delivered via caesarean section and who are formula fed as opposed to vaginally delivered and breast-fed have significantly lower gut populations of bifidobacteria.[9] The specific breakdown of bifidobacterial species varies but the major contributors include: B. brevi, B. longum, B. pseudocatenulatum, B. adolescentis, B. pseudolongum, and B. bifidum. The number of bifidobacteria in the fecal flora of adults, however, is much lower around 3-6%. Numerous studies have been conducted that illustrate a variance of gut microbiota both by subject as well as with subject age. B. bifidum is also found, although to a much lesser extent, in breast milk, the mouth, and the vagina.
Economic Importance
The numerous beneficial attributes of bifidobacteria have led to an increased awareness and the growing popularity of probiotics. Television commercials for Activia yogurt featuring the actress Jamie Lee Curtis have gained much notoriety. Probiotic supplements are relatively inexpensive and easily available to consumers both online and at local pharmacies without a prescription. The growing market for probiotics has sparked an influx of diverse product offerings, which include: yogurt, yogurt drinks, capsule supplements, cereal, snack bars, pediatric-specific supplements, baby formula, and toothpaste.
Current Research
Researchers have tried to understand and explain the correlation between the maternal gut microbiotic composition, the direct transfer to infant via breast-feeding, and the subsequent microbiotic composition of the infant gut. Two of the following studies (Effects... and Isolation...) have opposing views as to whether maternal gut composition and direct breast-feeding transfer method in fact influence infant gut composition.
Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity- a randomized, double-blind, placebo-controlled study.
The effects of prebiotics such as galactooligosaccharides (GOS) and long-chain fructooligosaccharides (lcFOS) that encourage the proliferation of bifidobacteria were examined on maternal gut microbiota during the third trimester of pregnancy and following delivery. This study aimed to determine whether maternal microbiota are passed to newborns via breast-feeding, and, as a secondary outcome, whether maternal gut microbiota affect the fetal immune response.[10] It is known that early infant gut microbiota influence an individual’s susceptibility to developing allergies later in life, although the exact reason and mechanism are unknown. The quantity of bifidobacteria and lactobacilli from the excised population was determined by fluorescent in situ hybridization and quantitative polymerase chain reaction in maternal and infant stool samples. The second objective, to examine the fetal immune response, utilized samples of cord blood by using flow cytometry and cytokine multiplex-array analysis. The results of this study relay that although GOS/lcFOS supplementation during pregnancy has a bifidogenic effect on maternal intestine micro flora, it is not passed on to newborns. Furthermore, there was no correlation between increased maternal bifidobacteria and enhanced fetal immune response.[11]
Exploring the diversity of the bifidobacterial population in the human intestinal tract.
While the beneficial aspects of bifidobacteria are widely accepted, the diversity and specific composition of the human intestine micro flora are less understood. Researchers conducted this study to identify specific bifidobacterial populations found in human intestines and fecal samples. Their protocol included plating human intestinal mucosal and fecal samples on selective media and further analyzing molecular data of selected rRNA gene sequences of individual colonies. Their findings clearly indicated that the majority, 704 of the 900 isolated colonies, were bifidobacteria. They further identified the six major species of bifidobacteria isolated from the intestine: B. longum, B. pseudocatenulatum, B. adolescentis, B. pseudolongum, B. breve, and B. bifidum, and two species found primarily in fecal samples, B. dentium and B. animalis subp. lactis. Their research indicated a correlation between age and the microbiota distribution of the intestine. A small selection of species were found exclusively in the adult human gut, while other species were found widely distributed. The study uncovered significant variance between individuals in the composition of fecal samples and intestinal mucosal samples as well as mild variance within the same subject (intrasubject variability) in different regions of the intestine. There were a small number of bifidobacteria that indicated the capacity to broadly colonize, which was based on the number that were able to be isolated from wide ecological distributions.[12]
Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR.
The aim of this study was to determine if breast milk contains bifidobacteria and further if it can be transferred from mother to infant via breast-feeding. This study examined 23 newborn infants that were exclusively breast-fed. Fructose-6-phosphate phosphoketolase assays were used to identify to the genus level and 16S rRNA gene sequencing to the species level. Bifidobacteria samples in breast milk were identified by PCR-denaturing gradient gel electrophoresis and approximate number determined by quantitative real-time PCR. Bifidobacteria were found in samples of both maternal milk and infant fecal matter. Eight of 23 mother child pairs showed a direct correlation between maternal milk microbial composition and infant feces microbial composition. Thus, this study concluded that breast milk serves as the source of bifidobacteria for the infant gut.[13]
References
- ↑ [1]
- ↑ Willey, J.M., Sherwood, L.M., Woolverton, C.J. Prescott, Harley, and Klein's Microbiology. Seventh edition. 2008
- ↑ Chen, X., Jiang, H., Yang, Y., Liu, N. “[Effect of exopolysaccharide from Bifidobacterium bifidum on cell of gastric cancer and human telomerase reverse transcriptase]”. Wei Sheng Wu Xue Bao. January, 2009. Vol. 49, Issue 1. p. 117-22
- ↑ [2]
- ↑ [3]
- ↑ Ventura, M., Canchaya, C., Tauch, A., Chandra, G., Fitzgerald, G.F., Chater, K.F., van Sinderen, D. “Genomics of Actinobacteria: Tracing the Evolutionary History of an Ancient Phylum”. Micobiol Mol Biol Rev. September, 2007. Vol. 71. p. 495-548
- ↑ Schell, M. A., Karmirantzou, M., Snel, B., Vilanova, D., Berger, B., Pessi, G., Zwahlen, M.C., Desiere, F., Bork, P., Delley, M., Pridmore, R.D., Arigoni, F. "The genome sequence of Bifidobacterium longum reflects its adaptation to the human gastrointestinal tract". Proc Natl Acad Sci. 2002. Vol. 99. p. 14422-14427
- ↑ Santacruz, A., Marcos, A., Warnberg, J., Marti, A., Martin-Matillas, M., Campoy, C., Moreno, L.A., Veiga, O., Redondo-Figuero, C., Garagorri, J.M., Azcona, C., Delgado, M., Garcia-Fuentes, M., Collado, M.C., Sanz, Y. “Interplay between weight loss and gut microbiota composition in overweight adolescents”. Obesity. April, 2009. doi:10.1038/oby.2009.112
- ↑ Gronlund, M.M., Gueimonde, M., Laitinen, K., Kociubinski, G., Gronroos, T., Salminen, S., Isolauri, E. “Maternal breast-milk and intestinal bifidobacteria guide the compositional development of the bifidobacterium microbiota in infants at risk of allergic disease”. Clinical and Experimental Allergy. August, 2007. Vol. 37. p. 1764-1772
- ↑ Mouni, F., Aissi, E., Hernandez, J., Gorocica, P., Bouquelet, S., Zenteno, E., Lascurain, R., Garfias, Y. “Effect of Bifidobacterium bifidum DSM 20082 cytoplasmic fraction on human immune cells”. Immunological Investigations. 2009. Vol. 38. p. 104-115
- ↑ Shadid, R., Haarman, M., Knol, J., Theis, W., Beermann, C., Rjosk-Dendorfer, D., Schendel, D., Koletzko, B., Krauss-Etschmann, S. “Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity-a randomized, double-blind, placebo-controlled study”. Am J Clin Nutr. 2009. Vol. 86. p. 1426-37
- ↑ Turroni, F., Foroni, E., Pizzetti, P., Giubellini, V., Ribbera, A., Merusi, P., Cagnasso, P., Bizzarri, B., de'Angelis, G.L., Shanahan, F., van Sinderen, D., Ventura, M. “Exploring the diversity of the bifidobacterial population in the human intestinal tract”. Appl Environ Microbiol. March, 2009. Vol. 75, Issue 6. p. 1534-45
- ↑ Martin, R., Jimenez, E., Heilig, H., Fernandez, L., Marin, M.L., Zoetendal, E.G., Rodriguez, J.M. “Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR”. Appl Environ Microbiol. February, 2009. Vol. 75, Issue 4. p. 965-9