Thermus aquaticus: Difference between revisions
imported>Golareh Sina |
mNo edit summary |
||
| (50 intermediate revisions by 4 users not shown) | |||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
| Line 5: | Line 4: | ||
| color = pink | | color = pink | ||
| name = Thermus aquaticus | | name = Thermus aquaticus | ||
| image = | | image = [[Image:Thermus aquaticus.JPG]] | ||
| regnum = Eubacteria | | regnum = Eubacteria | ||
| phylum = | | phylum = | ||
| Line 20: | Line 19: | ||
==Description and significance== | ==Description and significance== | ||
''Thermus aquaticus'' was isolated in 1969 by Brocks and Freeze of University of Indiana. It is a Gram-negative bacteria both motile (presence of a flagellum) or immotile. Comparisons between structures of ''T. aquaticus'' and ''E. coli'' have shown many similarities linking them to a common ancestor. YT-1 gene extracted from ''T. aquaticus'' is structurally similar to RecA from ''E. coli'' homologue and comparisons between Klentaq1, a large fragment of Taq DNA polymerase and the Klenow fragment of E Coli DNA polymerase reveal identical C-termini and very similar N-termini. | |||
Thermus | Discovery of this species of thermophiles primarily came as a surprise to researchers and very soon proved to be an extremely important tool in many fields of sciences including biology, microbiology, genetics, diagnostics, clinical laboratories, forensic and environmental sciences, hereditary studies and paternity testing. The importance of this discovery comes from the high thermostability of the ''T. aquaticus'' proteins. The taq polymerase plays an extremely important role in the polymerase chain reaction (PCR). PCR is a process by which one or a few stretches of DNA is amplified using thermal cycling introduced by Kary Mullis in 1984 winning him the Nobel Prize in 1993. The enzymes found in the ''T. aquaticus'' are able to withstand the heat in the denaturing of the newly formed DNA so strands can separate and act as templates for the next cycle of PCR. Taq polymerase, with an optimum activity at 72-80 degrees Celsius and a half life of 9 minutes at 97.5 degrees Celsius, can replicate 9000 base pairs in less than 10 seconds, and presently still plays an extremely important role in gaining insight into realm of biotechnology. Thermus Aquaticus has not been associated with any known pathology. | ||
==Genome structure== | ==Genome structure== | ||
''T. aquaticus'' has a double stranded circular DNA chromosome with a length of 2,338,193 nt., with a replicon type WGS, (Master Wgs), no pseudogenes, 53 structural RNA's, 1982 protein-coding sequences, and a pTT27 plasmid. | |||
==Cell structure and metabolism== | ==Cell structure and metabolism== | ||
"T. aquaticus" contains pillus like structures used in conjugation. Twelve genes in 3 loci were found to encode preplin-like proteins essential for natural transformation."T. aquaticus" not only can function at high temperatures but they thrive at elevated temperatures. Optimum growth is seen between 60 and 75°C (could go as low as 35°C to as high as 85°C). The optimal pH ranges from 7.5 to 8.0 generally, but some strains grow between pH 5.1 and 9.5. This organism is a chemotroph using | |||
carbohydrates, amino acids, caboxylic acids and peptides for growth. Monosaccharides such as glucose are generally used for carbons sources but sucrose, maltose could be used. (Icelandic stains). Some of the proteins isolated from this organism are: elastin, fibrin and casein. Not all strains can hydrolize all substrates. | |||
Membranes of these proteins are remarkably temperature stable and the heat stability of the enzymes and protein-synthesis systems allow them to function efficiently at high | |||
temperatures. Many factors contribute to the stability of the proteins: | |||
1) Highly organized hydrophobic interiors | |||
2) More hydrogen bonds and presence of other non-covalent bonds strengthen the protein structures. | |||
3) Larger quantities of amino acids like proline make peptide chains less flexible. | |||
4) Protein folding is aided and stabilized by special chaperone proteins. | |||
5) Some evidence indicates that DNA is stabilized by specialized histone-like proteins. | |||
6) Their membrane lipids tend to be more saturated and more branched and possess higher molecular weight resulting in a higher melting point and in turn more thermostability. | |||
==Ecology== | |||
''T. aquaticus'' was first isolated in the Great Fountain region of Yellowstone National Park from neutral and alkaline springs in 1969 by Brocks and Freeze. This discovery disproved the previous beliefs that bacteria could not function properly at higher temperatures. After this discovery, some strains of ''T. aquaticus'' were discovered in hot springs in Iceland and hydrothermal vents in other parts of the world. ''T. aquaticus'' is sometimes found living in conjunction with other organisms such as cyanobacteria and obtain some of the energy for their growth and metabolism from the neighboring bacterias' photosynthesis. | |||
==Application to Biotechnology== | |||
Enzymes derived from T. aquaticus have had an incredibly important role in facilitating many aspects of biotechnology correlated with DNA amplification. They enable researchers to study proteins and enzymes under conditions not possible before. This is all due to the thermostability of the proteins and their ability to function at even higher rates at high | |||
temperatures. Some of the isolated enzymes and their roles in facilitating applications in biotechnology are as follows: | |||
1) Adolase- | |||
2) RNA polymerase- first polymerase isolated from Taq in1974. | |||
3) Restriction Endonucleases | |||
4) DNA polymerase- isolated in 1976, could be isolated in purer form and later discovered to be used in PCR, for amplifying short segments of DNA (before the discovery of the Taq DNA, enzymes needed to be added after each cycle of denaturing of DNA, but with the use of the Taq DNA polymerase it was not necessary anymore.) One single copy of genomic sequence can be amplified by a factor of more than 10 million(One ng of DNA template up to 35 kb could be amplified from a target DNA molecule present only once in a sample of 105 cells) with high base pair fidelity. This enzyme was soon cloned, sequenced, and produced in mass quantities for commercial sale. | |||
5) Other enzymes with high optimal temperatures allowing researchers to study them in extreme conditions are:DNA ligase, alkaline phosphatase, NADH oxidase, isocitrase, dehydrogenase, amylomaltase and fructose1,6-bisphosphate-dependent L-lactate dehydrogenase. | |||
Besides the revolutionary changes in PCR, ligase chain reaction (LCR), which uses T. aquaticus ligase, can amplify genetic sequences of stretches of DNA that posses a desired sequence million or more times within hours. It can amplify and screen in a single step and screen for mutations simultaneously. LCR is useful in testing for hereditary diseases, revealing hidden infections and distinguishing between drug resistant and drug sensitive strains of viruses and bacteria. | |||
==Current Research== | |||
"THE GENOMOCS OF DISULFIDE BONDING AND PROTEIN STABILIZATION IN THERMOPHILES" | |||
Recent studies have emphasized the role of disulfide bonds in stabilizing the structure of intracellular proteins of Thermus Aquaticus among some other thermophiles. Previously the popular belief was disulfide bonds are only present in extracellular proteins where they stabilize folded proteins against harsh conditions and are rarely found in the cytosol. The specific protein which seems to be responsible for the formation of intracellular disulfide bonds seems to be protein disulfide oxidoreductase (PDO), which functions as a cytoplasmic protein disulfide-isomerase (PDI) . It has been suggested that eukaryotic PDI, found in the endoplasmic reticulum where it catalyzes isomerization of protein disulfide bonds, has evolved from a protein similar to thermophilic PDO. More research needs to be done on this subject. | |||
Besides thermophiles, elevated intracellular disulfide bonding has been seen in other extremophiles including; halophiles, alkylophiles, acidophiles, and radiation-tolerant organisms. This discovery supports the role of intracellular disulfide bonds in stabilizing proteins in all types of extreme conditions. This study sheds some light on different methods used by organisms to stabilize their proteins to adapt to "exotic" environments. <ref>Insert footnote text here</ref>(1) | |||
Controversy: After isolation of Thermus Aquaticus, samples of it were deposited in the American Type Cultures Collection (ATCC), a public repository. Other scientists had access to them and were able to do more research. By the 1980's, it became obvious that the potential for commercializing the enzymes from this species would prove to be very high and profitable. | |||
"Presence of Bacterial Phage-Like DNA Sequences in Commercial Taq DNA Polymerase Reagents, Tamara Newsome,1 Bing-Jie Li,1 Nianxiang Zou,1 and Shyh-Ching Lo2(2004)" | |||
In a routine study of amplifying the highly conserved gene sequence of the 16S rRNA, several DNA bands were produced that were different from the expected target sizes. Characteristics of these PCR products and further PCR study revealed they were amplified from commercial Taq DNA Polymerase reagents, apparently contaminated with trace amounts of bacteriophage like dnase. A study USING 16s rrna gene primer sA, a number of unexpected products with sizes ranging from 100bp more than 20 kb, in addition to expected 1.5KB. Site products or secondary products are not uncommon, however this time 3 PCR products, 3a, 3b, 7c, were further studied. They were gel purified, cloned, sequenced and compared to sequences in genbank database. A small portion of DNA sequence on 7C, showed 81% homology to tail fiber gene of pesudemona phage, GH-1 and 88% homology to tail fibers proteins of enterobacterial phage T7. 3A and 3B showed partial homology to tail tubular protein B(72%) and putative DNA ligase (55%) of pseudomonas phage GA-1. | |||
To confirm findings, four different primers were designed from phage like DNA sequences of these 3 bands and were studied by PCR to detect phage like DNA. The PCR band found were of expected sizes, so all PCR products were confirmed to be phage like DNA sequences by nucleotide sequencing. This experiment was repeated with Taq polymerases from two different commercial sources. PCR was performed against deionized water sample with or without UV treatment(which has been shown to reduce false positive signals). And the clone 3A, 3B and 7C DNA sequences were added into each group of reaction tubes to serve as positive control, no exogenous DNA template were added to any of the tubes. Hundred mM tris-HCL buffer with a pH 8.3200 micro molar deoxynucleoside triphosphates, 2.5mM MgCl2 25pmol of each primer and 1.25 Uof Taq polymerase. Phage like DNA found in two lots from company A and non from company B. Since company also provided MgCl2 solutions and buffers, they were also tested by swapping them with other samples of buffer and MgCl2. Still same results were observed. | |||
Presence of DNA contamination in Taq Polymerases were recorded before but most were exogenous bacterial DNA. This study is the first to show bacteriophage like DNA present. Scientist and Researchers were made aware of these findings. Using tainted amplification reagents can lead to misleading and confusing results. <ref>article name and arthur</ref> | |||
."PATENT RULING COULD CAUSE PCR ENZYME PRICES, Dalton, R (2001)" | |||
Hoffman La Roche, a Swiss-based pharmaceutical company, owns the patent rights for the Taq DNA polymerase enzyme and the National Parks system was not receiving any of the profits, even though the organism was discovered at a national park. The National Park Service called this "the great Taq ripoff". Since then, researchers in the national parks are required to sign an agreement of benefit sharing so a portion of the profits would be returned to the parks. Meanwhile, a fight for patent rights of the Taq polymerase is still going on. The European Patent office revoked Hoffman La Roche's patent claiming Taq Polymerase is naturally occurring and finding this enzyme was not a "novel invention". | |||
"EUROPEAN PATENT FOR PCR ENZYME CLOUDED BY RUSSIAN CLAIM, Dickson, D (1993)" | |||
Swiss pharmaceutical company acquired the patent rights for the Taq polymerase enzyme from the US Biotechnology company ; Cetus Corporation.While La Roche is fighting the European Patent Office to keep the patent rights, a Russian scientist named Stanislav Gorodetsky is claiming he and his research group were first to discover the organism "T. aquaticus" and isolate the enzyme and they should be involved in the profit sharing. Hoffman La Roche is insisting Russian enzyme is not identical. | |||
== | *{{Citation1- | ||
| title = 1-The Genomics of Disulfide Bonding and Protein Stabilization in Thermophiles | |||
| url http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=1188242 | |||
| title = The Genomics of Disulfide Bonding and Protein Stabilization in Thermophiles | |||
| year = 2005 | |||
| author = Beeby, M, O’Connor, B.D.,Ryttersgaard, C.,Boutz, D.R.,Perry, J., Yeates, T. | |||
| journal = PloS Biol. | |||
| pages = | |||
| volume = e309 | |||
| accessdate = 2009-05-12 | |||
}} | |||
==References== | ==References== | ||
| Line 80: | Line 106: | ||
Saiki, R.K.,et.al., Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science v.239 (January 29,1988)p.487-91. | Saiki, R.K.,et.al., Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science v.239 (January 29,1988)p.487-91. | ||
<http://microbewiki.kenyon.edu/index.php/Thermus> | |||
The Genomics of Disulfide Bonding and Protein Stabilization in Thermophiles | |||
Beeby M, O'Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, et al. | |||
PLoS Biology Vol. 3, No. 9, e309 doi:10.1371/journal.pbio.0030309 | |||
http://vnweb.hwwilsonweb.com.central.ezproxy.cuny.edu:2048/hww/jumpstart.jhtml?recid=0bc05f7a67b1790e5a5ef5947c67438bfc25e16bccc605e90e92bd260529e1f39f0fba67f43a661c&fmt=C | |||
Dalton, R. Patent ruling could cut PCR enzyme prices. Nature v. 411 no. 6838 (June 7 2001) p. 622 | |||
Dickson, D. European patent for PCR enzyme clouded by Russian claim. Nature v. 364 (July 1 1993) p. 2 | |||
[[Category:Suggestion Bot Tag]] | |||
Latest revision as of 06:01, 28 October 2024
| Thermus aquaticus | ||||||
|---|---|---|---|---|---|---|
[[image: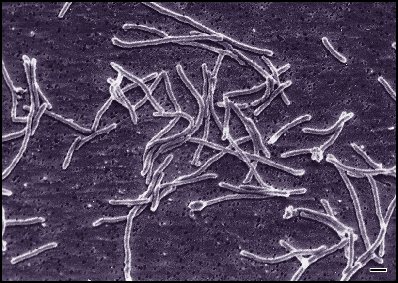 |200px|]] |200px|]] | ||||||
| Scientific classification | ||||||
|
Description and significance
Thermus aquaticus was isolated in 1969 by Brocks and Freeze of University of Indiana. It is a Gram-negative bacteria both motile (presence of a flagellum) or immotile. Comparisons between structures of T. aquaticus and E. coli have shown many similarities linking them to a common ancestor. YT-1 gene extracted from T. aquaticus is structurally similar to RecA from E. coli homologue and comparisons between Klentaq1, a large fragment of Taq DNA polymerase and the Klenow fragment of E Coli DNA polymerase reveal identical C-termini and very similar N-termini. Discovery of this species of thermophiles primarily came as a surprise to researchers and very soon proved to be an extremely important tool in many fields of sciences including biology, microbiology, genetics, diagnostics, clinical laboratories, forensic and environmental sciences, hereditary studies and paternity testing. The importance of this discovery comes from the high thermostability of the T. aquaticus proteins. The taq polymerase plays an extremely important role in the polymerase chain reaction (PCR). PCR is a process by which one or a few stretches of DNA is amplified using thermal cycling introduced by Kary Mullis in 1984 winning him the Nobel Prize in 1993. The enzymes found in the T. aquaticus are able to withstand the heat in the denaturing of the newly formed DNA so strands can separate and act as templates for the next cycle of PCR. Taq polymerase, with an optimum activity at 72-80 degrees Celsius and a half life of 9 minutes at 97.5 degrees Celsius, can replicate 9000 base pairs in less than 10 seconds, and presently still plays an extremely important role in gaining insight into realm of biotechnology. Thermus Aquaticus has not been associated with any known pathology.
Genome structure
T. aquaticus has a double stranded circular DNA chromosome with a length of 2,338,193 nt., with a replicon type WGS, (Master Wgs), no pseudogenes, 53 structural RNA's, 1982 protein-coding sequences, and a pTT27 plasmid.
Cell structure and metabolism
"T. aquaticus" contains pillus like structures used in conjugation. Twelve genes in 3 loci were found to encode preplin-like proteins essential for natural transformation."T. aquaticus" not only can function at high temperatures but they thrive at elevated temperatures. Optimum growth is seen between 60 and 75°C (could go as low as 35°C to as high as 85°C). The optimal pH ranges from 7.5 to 8.0 generally, but some strains grow between pH 5.1 and 9.5. This organism is a chemotroph using carbohydrates, amino acids, caboxylic acids and peptides for growth. Monosaccharides such as glucose are generally used for carbons sources but sucrose, maltose could be used. (Icelandic stains). Some of the proteins isolated from this organism are: elastin, fibrin and casein. Not all strains can hydrolize all substrates. Membranes of these proteins are remarkably temperature stable and the heat stability of the enzymes and protein-synthesis systems allow them to function efficiently at high temperatures. Many factors contribute to the stability of the proteins:
1) Highly organized hydrophobic interiors 2) More hydrogen bonds and presence of other non-covalent bonds strengthen the protein structures. 3) Larger quantities of amino acids like proline make peptide chains less flexible. 4) Protein folding is aided and stabilized by special chaperone proteins. 5) Some evidence indicates that DNA is stabilized by specialized histone-like proteins. 6) Their membrane lipids tend to be more saturated and more branched and possess higher molecular weight resulting in a higher melting point and in turn more thermostability.
Ecology
T. aquaticus was first isolated in the Great Fountain region of Yellowstone National Park from neutral and alkaline springs in 1969 by Brocks and Freeze. This discovery disproved the previous beliefs that bacteria could not function properly at higher temperatures. After this discovery, some strains of T. aquaticus were discovered in hot springs in Iceland and hydrothermal vents in other parts of the world. T. aquaticus is sometimes found living in conjunction with other organisms such as cyanobacteria and obtain some of the energy for their growth and metabolism from the neighboring bacterias' photosynthesis.
Application to Biotechnology
Enzymes derived from T. aquaticus have had an incredibly important role in facilitating many aspects of biotechnology correlated with DNA amplification. They enable researchers to study proteins and enzymes under conditions not possible before. This is all due to the thermostability of the proteins and their ability to function at even higher rates at high temperatures. Some of the isolated enzymes and their roles in facilitating applications in biotechnology are as follows:
1) Adolase- 2) RNA polymerase- first polymerase isolated from Taq in1974. 3) Restriction Endonucleases 4) DNA polymerase- isolated in 1976, could be isolated in purer form and later discovered to be used in PCR, for amplifying short segments of DNA (before the discovery of the Taq DNA, enzymes needed to be added after each cycle of denaturing of DNA, but with the use of the Taq DNA polymerase it was not necessary anymore.) One single copy of genomic sequence can be amplified by a factor of more than 10 million(One ng of DNA template up to 35 kb could be amplified from a target DNA molecule present only once in a sample of 105 cells) with high base pair fidelity. This enzyme was soon cloned, sequenced, and produced in mass quantities for commercial sale. 5) Other enzymes with high optimal temperatures allowing researchers to study them in extreme conditions are:DNA ligase, alkaline phosphatase, NADH oxidase, isocitrase, dehydrogenase, amylomaltase and fructose1,6-bisphosphate-dependent L-lactate dehydrogenase.
Besides the revolutionary changes in PCR, ligase chain reaction (LCR), which uses T. aquaticus ligase, can amplify genetic sequences of stretches of DNA that posses a desired sequence million or more times within hours. It can amplify and screen in a single step and screen for mutations simultaneously. LCR is useful in testing for hereditary diseases, revealing hidden infections and distinguishing between drug resistant and drug sensitive strains of viruses and bacteria.
Current Research
"THE GENOMOCS OF DISULFIDE BONDING AND PROTEIN STABILIZATION IN THERMOPHILES"
Recent studies have emphasized the role of disulfide bonds in stabilizing the structure of intracellular proteins of Thermus Aquaticus among some other thermophiles. Previously the popular belief was disulfide bonds are only present in extracellular proteins where they stabilize folded proteins against harsh conditions and are rarely found in the cytosol. The specific protein which seems to be responsible for the formation of intracellular disulfide bonds seems to be protein disulfide oxidoreductase (PDO), which functions as a cytoplasmic protein disulfide-isomerase (PDI) . It has been suggested that eukaryotic PDI, found in the endoplasmic reticulum where it catalyzes isomerization of protein disulfide bonds, has evolved from a protein similar to thermophilic PDO. More research needs to be done on this subject.
Besides thermophiles, elevated intracellular disulfide bonding has been seen in other extremophiles including; halophiles, alkylophiles, acidophiles, and radiation-tolerant organisms. This discovery supports the role of intracellular disulfide bonds in stabilizing proteins in all types of extreme conditions. This study sheds some light on different methods used by organisms to stabilize their proteins to adapt to "exotic" environments. [1](1)
Controversy: After isolation of Thermus Aquaticus, samples of it were deposited in the American Type Cultures Collection (ATCC), a public repository. Other scientists had access to them and were able to do more research. By the 1980's, it became obvious that the potential for commercializing the enzymes from this species would prove to be very high and profitable.
"Presence of Bacterial Phage-Like DNA Sequences in Commercial Taq DNA Polymerase Reagents, Tamara Newsome,1 Bing-Jie Li,1 Nianxiang Zou,1 and Shyh-Ching Lo2(2004)"
In a routine study of amplifying the highly conserved gene sequence of the 16S rRNA, several DNA bands were produced that were different from the expected target sizes. Characteristics of these PCR products and further PCR study revealed they were amplified from commercial Taq DNA Polymerase reagents, apparently contaminated with trace amounts of bacteriophage like dnase. A study USING 16s rrna gene primer sA, a number of unexpected products with sizes ranging from 100bp more than 20 kb, in addition to expected 1.5KB. Site products or secondary products are not uncommon, however this time 3 PCR products, 3a, 3b, 7c, were further studied. They were gel purified, cloned, sequenced and compared to sequences in genbank database. A small portion of DNA sequence on 7C, showed 81% homology to tail fiber gene of pesudemona phage, GH-1 and 88% homology to tail fibers proteins of enterobacterial phage T7. 3A and 3B showed partial homology to tail tubular protein B(72%) and putative DNA ligase (55%) of pseudomonas phage GA-1. To confirm findings, four different primers were designed from phage like DNA sequences of these 3 bands and were studied by PCR to detect phage like DNA. The PCR band found were of expected sizes, so all PCR products were confirmed to be phage like DNA sequences by nucleotide sequencing. This experiment was repeated with Taq polymerases from two different commercial sources. PCR was performed against deionized water sample with or without UV treatment(which has been shown to reduce false positive signals). And the clone 3A, 3B and 7C DNA sequences were added into each group of reaction tubes to serve as positive control, no exogenous DNA template were added to any of the tubes. Hundred mM tris-HCL buffer with a pH 8.3200 micro molar deoxynucleoside triphosphates, 2.5mM MgCl2 25pmol of each primer and 1.25 Uof Taq polymerase. Phage like DNA found in two lots from company A and non from company B. Since company also provided MgCl2 solutions and buffers, they were also tested by swapping them with other samples of buffer and MgCl2. Still same results were observed. Presence of DNA contamination in Taq Polymerases were recorded before but most were exogenous bacterial DNA. This study is the first to show bacteriophage like DNA present. Scientist and Researchers were made aware of these findings. Using tainted amplification reagents can lead to misleading and confusing results. [2]
."PATENT RULING COULD CAUSE PCR ENZYME PRICES, Dalton, R (2001)"
Hoffman La Roche, a Swiss-based pharmaceutical company, owns the patent rights for the Taq DNA polymerase enzyme and the National Parks system was not receiving any of the profits, even though the organism was discovered at a national park. The National Park Service called this "the great Taq ripoff". Since then, researchers in the national parks are required to sign an agreement of benefit sharing so a portion of the profits would be returned to the parks. Meanwhile, a fight for patent rights of the Taq polymerase is still going on. The European Patent office revoked Hoffman La Roche's patent claiming Taq Polymerase is naturally occurring and finding this enzyme was not a "novel invention".
"EUROPEAN PATENT FOR PCR ENZYME CLOUDED BY RUSSIAN CLAIM, Dickson, D (1993)"
Swiss pharmaceutical company acquired the patent rights for the Taq polymerase enzyme from the US Biotechnology company ; Cetus Corporation.While La Roche is fighting the European Patent Office to keep the patent rights, a Russian scientist named Stanislav Gorodetsky is claiming he and his research group were first to discover the organism "T. aquaticus" and isolate the enzyme and they should be involved in the profit sharing. Hoffman La Roche is insisting Russian enzyme is not identical.
References
Barnes,W.M. PCR amplification of up to 35 kb DNA with high fidelity and high yield from bacteriophage templates-proceedings of the National Academy of Sciences of the United States of America v.91(March 15,1994)p.2216-20
Weiss, R. Hot prospect for new gene amplifier-Science v.254(November 29,1991)p.1292-3
Saiki, R.K.,et.al., Primer directed enzymatic amplification of DNA with a thermostable DNA polymerase. Science v.239 (January 29,1988)p.487-91.
<http://microbewiki.kenyon.edu/index.php/Thermus> The Genomics of Disulfide Bonding and Protein Stabilization in Thermophiles Beeby M, O'Connor BD, Ryttersgaard C, Boutz DR, Perry LJ, et al. PLoS Biology Vol. 3, No. 9, e309 doi:10.1371/journal.pbio.0030309
Dalton, R. Patent ruling could cut PCR enzyme prices. Nature v. 411 no. 6838 (June 7 2001) p. 622
Dickson, D. European patent for PCR enzyme clouded by Russian claim. Nature v. 364 (July 1 1993) p. 2