Single-nucleotide polymorphism: Difference between revisions
imported>Daniel Mietchen (started) |
imported>Daniel Mietchen (brought over from WP) |
||
| Line 1: | Line 1: | ||
{{subpages}} | {{subpages}} | ||
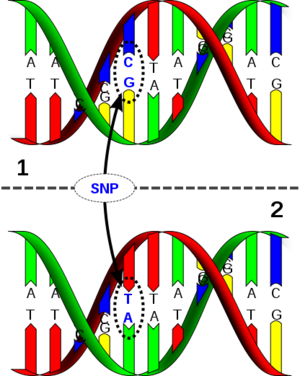
{{Image|416px-Dna-SNP.svg.png|thumb|DNA molecule 1 differs from DNA molecule 2 at a single base-pair location (a C/T polymorphism).}} | |||
A '''single-nucleotide polymorphism''' ('''SNP''', pronounced ''snip'') is a [[DNA sequence]] variation occurring when a single [[nucleotide]] — [[adenine|A]], [[thymine|T]], [[cytosine|C]], or [[guanine|G]] — in the [[genome]] (or other shared sequence) differs between members of a species (or between paired chromosomes in an individual). For example, two sequenced DNA fragments from different individuals, AAGC'''C'''TA to AAGC'''T'''TA, contain a difference in a single nucleotide. In this case we say that there are two ''[[allele]]s'' : C and T. Almost all common SNPs have only two alleles. | |||
Within a population, SNPs can be assigned a [[minor allele frequency]] — the lowest allele frequency at a [[locus (genetics)|locus]] that is observed in a particular population. | |||
This is simply the lesser of the two allele frequencies for single-nucleotide polymorphisms[1]. There are variations between human populations, so a SNP allele that is common in one geographical or ethnic group may be much rarer in another. | |||
== "SNP" == | |||
In the past, SNPs with a minor allele frequency of greater than or equal to 1% (or 0.5%, etc.) were given the title "SNP".<ref>E.g., {{Cite web | |||
| title = Methods for Discovering and Scoring Single Nucleotide Polymorphisms | |||
| url = http://www.genome.gov/10001029 | |||
| publisher = [[National Human Genome Research Institute]] | |||
}}</ref> | |||
Some used "[[mutation]]" to refer to variations with low allele frequency. | |||
With the advent of modern [[bioinformatics]] and a better understanding of evolution, this definition is no longer necessary, e.g., a database such as [[dbSNP]] includes "SNPs" that have lower allele frequency than one percent.<ref>{{Cite news | |||
| url = http://www.ncbi.nlm.nih.gov/Web/Newsltr/Summer02/snp.html | |||
| title = SNP Population Grows at NCBI | |||
| publisher = NCBI | |||
| work = NCBI News | |||
}}</ref> | |||
== Types of SNPs == | |||
{| class="infobox" | |||
|- | |||
! Types of SNPs | |||
|- | |||
| | |||
* Non-coding region | |||
* Coding region | |||
** Synonymous | |||
** Nonsynonymous | |||
*** Missense | |||
*** Nonsense | |||
|} | |||
Single-nucleotide [[polymorphism (biology)|polymorphisms]] may fall within coding sequences of [[gene]]s, [[intron|non-coding regions of genes]], or in the [[intergenic region]]s between genes. | |||
SNPs within a coding sequence will not necessarily change the [[amino acid]] sequence of the [[protein]] that is produced, due to [[Genetic code#Degeneracy of the genetic code|degeneracy of the genetic code]]. | |||
A SNP in which both forms lead to the same polypeptide sequence is termed ''[[synonymous mutation|synonymous]]'' (sometimes called a [[silent mutation]]) — if a different polypeptide sequence is produced they are ''[[nonsynonymous mutation|nonsynonymous]]''. | |||
A nonsynonymous change may either be [[missense mutation|missense]] or "[[nonsense mutation|nonsense]]", where a missense change results in a different amino acid, while a nonsense change results in a premature [[stop codon]]. | |||
SNPs that are not in protein-coding regions may still have consequences for [[gene splicing]], [[transcription factor]] binding, or the sequence of [[non-coding RNA]]. | |||
== Use and importance of SNPs == | |||
Variations in the DNA sequences of humans can affect how humans develop [[disease]]s and respond to [[pathogen]]s, [[chemical]]s, [[medication|drugs]], [[vaccine]]s, and other agents. SNPs are also thought to be key enablers in realizing the concept of [[personalized medicine]].<ref>{{cite news | author=Bruce Carlson | title=SNPs — A Shortcut to Personalized Medicine | |||
| url=http://www.genengnews.com/articles/chitem.aspx?aid=2507 | work=[[Genetic Engineering & Biotechnology News]] | |||
| publisher=[[Mary Ann Liebert, Inc.]] | page=12 | date=2008-06-15 | accessdate=2008-07-06 | |||
| quote=(subtitle) Medical applications are where the market's growth is expected }}</ref> However, their greatest importance in biomedical research is for comparing regions of the genome between [[cohorts]] (such as with matched cohorts with and without a disease). | |||
The study of single-nucleotide polymorphisms is also important in crop and livestock breeding programs (see [[genotyping]]). See [[SNP genotyping]] for details on the various methods used to identify SNPs. | |||
== Examples == | |||
Example SNPs are [[rs6311]] and [[rs6313]] in the [[HTR2A]] gene. | |||
A SNP in the ''[[F5 (gene)|F5]]'' gene causes a hypercoagulability disorder with the variant [[Factor V Leiden]]. | |||
An example of a triallelic SNP is [[rs3091244]].<ref>{{Cite journal | |||
| author = Akihiko Morita, Tomohiro Nakayama, Nobutaka Dobac, Shigeaki Hinoharac, Tomohiko Mizutania and Masayoshi Soma | |||
| title = Genotyping of triallelic SNPs using TaqMan® PCR | |||
| journal = Molecular and Cellular Probes | |||
| volume = 21 | |||
| issue = 3 | |||
| month = june | |||
| year = 2007 | |||
| pages = 171–176 | |||
}}</ref> | |||
== Databases == | |||
As there are for genes, there are also [[bioinformatics]] databases for SNPs. | |||
''[[dbSNP]]'' is a SNP database from [[National Center for Biotechnology Information]] (NCBI). | |||
''[[SNPedia]]'' is a wiki-style database from a private company. | |||
The ''[[OMIM]]'' database describes the association between polymorphisms and, e.g., diseases. | |||
== Nomenclature == | |||
The nomenclature for SNPs can be confusing: several variations can exist for an individual SNP and consensus has not yet been achieved. One approach is to write SNPs with a prefix, period and greater than sign showing the wild-type and altered nucleotide or amino acid; for example, c.76A>T.<ref>{{Cite web | |||
| author = J.T. Den Dunnen | |||
| title = Recommendations for the description of sequence variants | |||
| date = 2008-02-20 | |||
| accessdate = 2008-09-05 | |||
| url = http://www.hgvs.org/mutnomen/recs.html | |||
| publisher = [[Human Genome Variation Society]] | |||
}}</ref><ref>{{Cite journal | |||
| author = Johan T. den Dunnen & Stylianos E. Antonarakis | |||
| title = Mutation Nomenclature Extensions and Suggestions to Describe Complex Mutations: A Discussion | |||
| journal = [[Human Mutation]] | |||
| volume = 15 | |||
| pages = 7–12 | |||
| year = 2000 | |||
| url = http://www3.interscience.wiley.com/cgi-bin/fulltext/68503056/PDFSTART | |||
}}</ref><ref>{{Cite journal | |||
| author = Shuji Ogino, Margaret L. Gulley, Johan T. den Dunnen, Robert B. Wilson and the Association for Molecular Pathology Training and Education Committee | |||
| title = Standard Mutation Nomenclature in Molecular Diagnostics | |||
| journal = The Journal of Molecular Diagnostics | |||
| year = 2007 | |||
| volume = 9 | |||
| issue= 1 | |||
| doi = 10.2353/jmoldx.2007.060081 | |||
}}</ref> | |||
== See also == | |||
*[[Single-base extension]] | |||
* [[Variome]] | |||
* [[TaqMan]] | |||
* [[Affymetrix]] | |||
* [[International HapMap Project]] | |||
* [[tag SNP]] | |||
==Notes== | |||
{{reflist}} | |||
==References== | |||
*[http://www.nature.com/nrg/journal/v5/n2/glossary/nrg1270_glossary.html] | |||
*[http://www.ornl.gov/sci/techresources/Human_Genome/faq/snps.shtml Human Genome Project Information] — SNP Fact Sheet | |||
*[http://nci.nih.gov/cancertopics/understandingcancer/geneticvariation/Slide1 Relation of SNP's with Cancer] | |||
==External links== | |||
*[http://www.ncbi.nlm.nih.gov/About/primer/snps.html NCBI resources] — Introduction to SNPs from NCBI | |||
*[http://snp.cshl.org/ The SNP Consortium LTD] — SNP search | |||
*[http://www.ncbi.nlm.nih.gov/projects/SNP/ NCBI dbSNP database] — "a central repository for both single base nucleotide substitutions and short deletion and insertion polymorphisms" | |||
*[http://hapmap.org/ International HapMap Project] — "a public resource that will help researchers find genes associated with human disease and response to pharmaceuticals" | |||
*[http://www.1000genomes.org/ 1000 Genomes Project] — A Deep Catalog of Human Genetic Variation | |||
*[http://www.glovar.org/ Glovar Variation Browser] — variation information in a genomic context | |||
*[http://sift.jcvi.org SIFT] — "An online tool that predicts on the effect of SNPs on protein function" | |||
*[http://watcut.uwaterloo.ca/watcut/watcut/template.php WatCut] — an online tool for the design of SNP-RFLP assays | |||
*[http://bioinfo.iconcologia.net/index.php?module=Snpstats SNPStats] — SNPStats, a web tool for analysis of genetic association studies | |||
*[http://insilico.ehu.es/restriction Restriction HomePage] — a set of tools for DNA restriction and SNP detection, including design of mutagenic primers | |||
*[http://www.aacr.org/home/public--media/for-the-media/fact-sheets/cancer-concepts/snps.aspx American Association for Cancer Research Cancer Concepts Factsheet on SNPs] | |||
*[http://www.pharmgkb.org PharmGKB] — The Pharmacogenetics and Pharmacogenomics Knowledge Base, a resource for SNPs associated with drug response and disease outcomes. | |||
*[http://www.argusbio.com/sooryakiran/gensnip/gensnip.php GEN-SNiP] — Online tool that identifies polymorphisms in test DNA sequences. | |||
*[http://sift.jcvi.org Online tool that predicts on the effects of SNPs on protein function] | |||
*[http://www.informatics.jax.org/mgihome/nomen/gene.shtml Rules for Nomenclature of Genes, Genetic Markers, Alleles, and Mutations in Mouse and Rat] | |||
*[http://www.gene.ucl.ac.uk/nomenclature/guidelines.html HGNC Guidelines for Human Gene Nomenclature] | |||
Revision as of 11:20, 29 January 2009
A single-nucleotide polymorphism (SNP, pronounced snip) is a DNA sequence variation occurring when a single nucleotide — A, T, C, or G — in the genome (or other shared sequence) differs between members of a species (or between paired chromosomes in an individual). For example, two sequenced DNA fragments from different individuals, AAGCCTA to AAGCTTA, contain a difference in a single nucleotide. In this case we say that there are two alleles : C and T. Almost all common SNPs have only two alleles.
Within a population, SNPs can be assigned a minor allele frequency — the lowest allele frequency at a locus that is observed in a particular population. This is simply the lesser of the two allele frequencies for single-nucleotide polymorphisms[1]. There are variations between human populations, so a SNP allele that is common in one geographical or ethnic group may be much rarer in another.
"SNP"
In the past, SNPs with a minor allele frequency of greater than or equal to 1% (or 0.5%, etc.) were given the title "SNP".[1] Some used "mutation" to refer to variations with low allele frequency. With the advent of modern bioinformatics and a better understanding of evolution, this definition is no longer necessary, e.g., a database such as dbSNP includes "SNPs" that have lower allele frequency than one percent.[2]
Types of SNPs
| Types of SNPs |
|---|
|
Single-nucleotide polymorphisms may fall within coding sequences of genes, non-coding regions of genes, or in the intergenic regions between genes. SNPs within a coding sequence will not necessarily change the amino acid sequence of the protein that is produced, due to degeneracy of the genetic code. A SNP in which both forms lead to the same polypeptide sequence is termed synonymous (sometimes called a silent mutation) — if a different polypeptide sequence is produced they are nonsynonymous. A nonsynonymous change may either be missense or "nonsense", where a missense change results in a different amino acid, while a nonsense change results in a premature stop codon. SNPs that are not in protein-coding regions may still have consequences for gene splicing, transcription factor binding, or the sequence of non-coding RNA.
Use and importance of SNPs
Variations in the DNA sequences of humans can affect how humans develop diseases and respond to pathogens, chemicals, drugs, vaccines, and other agents. SNPs are also thought to be key enablers in realizing the concept of personalized medicine.[3] However, their greatest importance in biomedical research is for comparing regions of the genome between cohorts (such as with matched cohorts with and without a disease).
The study of single-nucleotide polymorphisms is also important in crop and livestock breeding programs (see genotyping). See SNP genotyping for details on the various methods used to identify SNPs.
Examples
Example SNPs are rs6311 and rs6313 in the HTR2A gene. A SNP in the F5 gene causes a hypercoagulability disorder with the variant Factor V Leiden. An example of a triallelic SNP is rs3091244.[4]
Databases
As there are for genes, there are also bioinformatics databases for SNPs. dbSNP is a SNP database from National Center for Biotechnology Information (NCBI). SNPedia is a wiki-style database from a private company. The OMIM database describes the association between polymorphisms and, e.g., diseases.
Nomenclature
The nomenclature for SNPs can be confusing: several variations can exist for an individual SNP and consensus has not yet been achieved. One approach is to write SNPs with a prefix, period and greater than sign showing the wild-type and altered nucleotide or amino acid; for example, c.76A>T.[5][6][7]
See also
Notes
- ↑ E.g., Methods for Discovering and Scoring Single Nucleotide Polymorphisms. National Human Genome Research Institute.
- ↑ SNP Population Grows at NCBI, NCBI News, NCBI.
- ↑ Bruce Carlson. SNPs — A Shortcut to Personalized Medicine, Genetic Engineering & Biotechnology News, Mary Ann Liebert, Inc., 2008-06-15, p. 12. Retrieved on 2008-07-06. “(subtitle) Medical applications are where the market's growth is expected”
- ↑ Akihiko Morita, Tomohiro Nakayama, Nobutaka Dobac, Shigeaki Hinoharac, Tomohiko Mizutania and Masayoshi Soma (june 2007). "Genotyping of triallelic SNPs using TaqMan® PCR". Molecular and Cellular Probes 21 (3): 171–176.
- ↑ J.T. Den Dunnen (2008-02-20). Recommendations for the description of sequence variants. Human Genome Variation Society. Retrieved on 2008-09-05.
- ↑ Johan T. den Dunnen & Stylianos E. Antonarakis (2000). "Mutation Nomenclature Extensions and Suggestions to Describe Complex Mutations: A Discussion". Human Mutation 15: 7–12.
- ↑ Shuji Ogino, Margaret L. Gulley, Johan T. den Dunnen, Robert B. Wilson and the Association for Molecular Pathology Training and Education Committee (2007). "Standard Mutation Nomenclature in Molecular Diagnostics". The Journal of Molecular Diagnostics 9 (1). DOI:10.2353/jmoldx.2007.060081. Research Blogging.
References
- [1]
- Human Genome Project Information — SNP Fact Sheet
- Relation of SNP's with Cancer
External links
- NCBI resources — Introduction to SNPs from NCBI
- The SNP Consortium LTD — SNP search
- NCBI dbSNP database — "a central repository for both single base nucleotide substitutions and short deletion and insertion polymorphisms"
- International HapMap Project — "a public resource that will help researchers find genes associated with human disease and response to pharmaceuticals"
- 1000 Genomes Project — A Deep Catalog of Human Genetic Variation
- Glovar Variation Browser — variation information in a genomic context
- SIFT — "An online tool that predicts on the effect of SNPs on protein function"
- WatCut — an online tool for the design of SNP-RFLP assays
- SNPStats — SNPStats, a web tool for analysis of genetic association studies
- Restriction HomePage — a set of tools for DNA restriction and SNP detection, including design of mutagenic primers
- American Association for Cancer Research Cancer Concepts Factsheet on SNPs
- PharmGKB — The Pharmacogenetics and Pharmacogenomics Knowledge Base, a resource for SNPs associated with drug response and disease outcomes.
- GEN-SNiP — Online tool that identifies polymorphisms in test DNA sequences.
- Online tool that predicts on the effects of SNPs on protein function
- Rules for Nomenclature of Genes, Genetic Markers, Alleles, and Mutations in Mouse and Rat
- HGNC Guidelines for Human Gene Nomenclature