Systems biology: Difference between revisions
imported>Anthony.Sebastian |
imported>Anthony.Sebastian (Adding recommende book) |
||
| Line 381: | Line 381: | ||
=== Books=== | === Books=== | ||
*Boogerd FC, Bruggeman FJ, Hofmeyr J-HS, Westerhoff HV (editors) (2007) Systems Biology: Philosophical Foundations. Elsevier ISBN 0444520856 | |||
:: 360 pages; 14 chapters; [http://books.elsevier.com/us//pharmatox/us/subindex.asp?maintarget=&isbn=9780444520852&country=United+States&srccode=&ref=goog&subcode=&head=&pdf=&basiccode=&txtSearch=&SearchField=&operator=&order=&community=pharmatox Book description and table of contents] | |||
*Alon U. ('''2007''') ''An Introduction to Systems Biology: Design Principles of Biological Circuits''. Boca Raton: Chapman and Hall/CRC ISBN 1-58488-642-0 | *Alon U. ('''2007''') ''An Introduction to Systems Biology: Design Principles of Biological Circuits''. Boca Raton: Chapman and Hall/CRC ISBN 1-58488-642-0 | ||
::301 pages; 12 chapters; 4 appendixes; 23 pages of references; glossary; [http://search.barnesandnoble.com/booksearch/isbninquiry.asp?ean=9781584886426&displayonly=TOC&z=y#TOC/ Table of Contents] | ::301 pages; 12 chapters; 4 appendixes; 23 pages of references; glossary; [http://search.barnesandnoble.com/booksearch/isbninquiry.asp?ean=9781584886426&displayonly=TOC&z=y#TOC/ Table of Contents] | ||
Revision as of 21:19, 22 July 2007
|
As an academic discipline, systems biology aims to explain, predict and control the properties, functions and behaviors[1] of biological systems — complex assemblages of interrelated, dynamically interacting, coordinated and hierarchically organized naturally selected components[2] generated over evolutionary time by natural biochemical and biological experiments.[3] Biological systems studied range from the level of the molecular subsystems of cells (e.g., mitochondrial production of versatilely usable energy; circuits regulating gene expression; cell-to-cell and within-cell signaling (communicational) pathways); to the Earth's biosphere, and include a large number of intermediate system levels (e.g., unicellular organisms, multicellular organisms, ecosystems).
Systems biology developed as a scientific endeavor in part because of the recognition that knowledge of the properties of a system’s components separated from the system embedding them, however complete, could not yield an explanation of the organized behavior of the system, which depends not only on the properties of the system’s components, but also on how they interrelate and interact in the dynamically and coordinately spatio-temporally, and on how the organized behavior of the system, influenced in part by the larger system embedding it, itself influences the properties and interactions of the components. Novel and unpredictable properties of systems emerge from its dynamic organization, the understanding of which requires a ‘holistic’ [4] approach in addition to the traditional ‘reductionistic’[4] approach. (See text-box at right.)
Systems biologists try to achieve their aims in part through developing models (descriptions, representations, simulations) of systems — various types of models, including graphical, mathematical and computer-based simulation models. The models aspire to 'describe' systems, or 'translate' them into the known 'language' of the model, in order to enable interpretation of the systems' components' organization and the systems' behavior by human cognition. The models aspire to do that sufficiently well to account for experimentally derived data about a system’s coordinated dynamically interacting components, and allow predictions of the system's properties, functions and behaviors in response to given stimuli or given sets of conditions. Systems biologists use sophisticated mathematical, statistical and computational tools in diverse modeling approaches, in mathematical network analyses, in computer simulations,[5] in design and building synthetic networks from biomolecules, and, iteratively, incorporating new data derived from systems-analysis-inspired further experimentation for the purpose of refining their models.
Mathematical modeling permits a kind of formal integrative analysis of a biological system that also facilitates the development of conceptual frameworks to integrate large amounts of data and identify gaps in the information known about a system. Without models, the human mind can comprehend little of the organization as a whole of a complex living system. In speaking specifically about the subsystem of cellular signaling pathways, Weng et al. put it this way: There is simply too much essential detail in biological signaling for the unaided human mind to organize and understand.[6]
Systems biologists often work with non-biological scientists, from a variety of disciplines, in developing models and conceptual frameworks. Some of those non-biological disciplines (e.g., mathematics, physics and chemistry) have developed systems approaches to explore systems in their own domains.
No definition or succinct description can capture the breadth and depth of the interdisciplinary enterprise of systems biology. Indeed, historian and philosopher of science Evelyn Fox Keller argues that “so far, ‘systems biology’ is a concept waiting for definition".[7] This article elaborates on the above description of the discipline.
Methodologies in general terms
Systems biologists try to accomplish their aims in part:
- by identifying and quantifying the system's components — for cells, acquiring so-called '-omics' data, including the numbers and types of nucleic acids, proteins, and metabolites;
- by identifying, characterizing, and to the extent possible, quantifying the interrelations (structural) and interactions (dynamical, coordinated, hierarchical) among the components of a system; for example, (a) for molecular components: gene and protein interactions involved in a cell’s metabolic pathways; (b) for organism components: organ interactions resulting in blood pressure homeostasis; for ecosystem components: predator and prey behaviors;
- by attempting to organize the system’s components abstractly in the form of 'models'/'simulations' — typically mathematical and computational — which encode such concepts as 'modules', 'circuits' and 'networks', 'network motifs', 'feedback' and 'feedforward' loops, 'homeostasis', 'adaptability', hierarchies, 'robustness, 'complexity', 'adaptation', and 'emergence';
- by conducting additional experiments to define interrelations and interactions of the real system that the model/simulation highlighted as knowledge gaps;
- by refining the model/simulation on the basis of the data from those model-inspired experiments;
- by designing, constructing and testing synthetic biological systems from biological components;[8]
- by developing new experimental technologies and new theoretical methods as required to answer new questions that inevitably arise in the study the unfathomable complexity of living systems.[9].
Modeling may take either so-called ‘bottom-up’ or ‘top-down’ approaches.[10] In bottom-up approaches, biologists take as their starting point the properties of molecules, or of elemental subsystems, and use that data to design models that explain the properties of the system that embeds them. They then evaluate the model’s predictions through experiments on the real system, and, depending on the results, they adjust the model and repeat the process, or they try to obtain additional experimental data that seem needed for a better explanatory/predictive model.
In top-down approaches, biologists take as their starting point the properties, functions and behaviors of the system-as-a-whole (system-data), and try to design models of component composition and interactions that both accord with the experimentally-derived component/interaction-data and account for the system-data. Often the system-data includes the responses of the system to typical perturbations. Deficiencies of the model’s accordance and accounting may suggest possibilities for the kinds of component-composition and interactions needed to make the model work, leading to targeted experimentation to discover new molecular or subsystem mechanisms operating in the system. As the model becomes refined through iterations of model-adjustment and targeted experimentation, it inspires better and better testable hypotheses about the system’s organization, potentially leading to both a complete description of the component-composition and its organizational dynamics and functionality. However, the complete data complement of the system often greatly outstrips the sometimes enormous data-sets available, or technologically acquirable, such that the most refined models offer only a glimpse of the system’s complexity.
For some systems, modeling may require the use of multiple datasets distributed in cyberspace, more than one mathematical or computational methodology, and even an array of interlinked computers having different architectures.[11] Many of the books and articles listed for further reading at the end of this article provide detailed descriptions and examples of the vast array of specific mathematical, graphical, computational, and biostatistical tools that systems biologists use to study biological systems.
Systems biologists expect progress in the field to yield explanations of biological systems suitable for applications in ecology, ethology, medicine, agriculture, business, the environment and technology — and to a considerable extent it has already done so. Some systems biologists consider the discipline critical to further progress in biology.[12]
The study of biological systems has expanded greatly as the result of major advances in molecular biology and data processing/analysis in the late 20th and early 21st century. Those include sequencing of genomes, developments in genetic engineering, developments of technologies for generating massive amounts of data on the structure and interactions of cellular components, and developments in bioinformatics &mdsh; all fueled by increasing interest by non-biological scientists (mathematicians, statisticians, computer scientists, physicists, chemists, and engineers, among others) in applying the principles and methods of their disciplines to the explanation of the improbable adaptive complexity of living systems (see Figure 1).
On the nature of biological "systems"
A 'system' in biology is any interconnected, interacting, coordinated and hierarchical assembly of biological components or elements—an organized assemblage with an identifiable function. For example, the vertebrate body is an assembly of diverse, interacting organs, among other components, functioning to survive and reproduce. See other examples below. Each component in a biological system interacts in some way(s) with one or more components in the system--a dynamical assembly of components. For example, in the system constituting a cell, proteins are the products of genes, but they also interact with genes, affecting their expression, as well as with other proteins. Systems can exhibit behaviors that are characteristic of the system-as-a-whole (see below), but which are not shared by any of its components (so-called emergent behaviors). A tree fruits, for example, because its dynamically interacting components enable it to, but no single component of a tree can. (More on emergent behavior below.)
Subsystems consist of smaller (less complex) systems embedded in a larger (more complex) system, and constitute at least part of the components or elements of the larger system. Whether a systems biologist treats a given assembly of components as a subsystem or as a system depends on the 'level' at which she focuses her attention. If she focuses her research at the level of a whole vertebrate organism, for example, she treats its organs as subsystems. If she focuses her research at the level of the heart, she treats the heart's interacting components as a system, while recognizing that the heart remains a component or element of a larger system (the circulatory system).
Even the larger systems, such as the vertebrate body system, are components or elements of even larger systems, a species of vertebrates, say, where individual members of the species interact with each other, as components, to generate a set of behaviors or properties characteristic of the species but not of the individual members of the species. The flocking behavior of birds illustrates a species behavior--technically the behavior of a deme. One bird cannot flock.
When trying to understand biological systems, systems biologists need not treat the components or elements of a system (or subsystem) exclusively as discrete or concrete objects or entities (e.g., molecules, organelles, cells, etc.), but may also treat them as abstracted concepts of organizational collections or activity patterns of those objects or entities (e.g., networks), which admit of study by graphical, mathematical, computational and statistical tools. Those include such concepts as circuits, networks and modules, more about which will follow below. Such concepts have a way of appearing less abstract or hypothetical as biologists more fully define them in terms of structure and coordinated dynamical interactions; predict systems behavior from them using quantitative models; and relate them functionally in the larger systems embedding them. Indeed, Bruggeman and Westerhoff[13] remark that the advances in molecular biology and mathematical modeling have "...shifted the focus of research from molecules to networks...", citing Barabasi and Otvai's review[14] underscoring the importance of networks in understanding the organization of cells.
Networks
Networks[15] ‘re-present’ or 'abstract' a system as an assemblage of 'nodes’ and ‘interactions’ among the nodes (also referred to as ‘edges’ or ‘arrows’ or ‘links’). In a spoken sentence, for example, words and phrases make up the nodes, and the interconnections of syntax (subject-to-predicate, preposition-to-object of preposition, etc.) make up the links. Molecular networks in cells represent specific functions in the cell; molecules make up the nodes, and their interactions with other nodes make up the edges or arrows. Networks accept inputs of one kind and return outputs of a different kind.
One finds networks everywhere in biology, from intracellular signaling pathways, to intraspecies networks, to ecosystems. Humans deliberately construct social networks of individuals working (more or less) to a common purpose, such as the U.S. Congress; they also construct networks of electronic parts to produce, for example, mobile phones; and networks of sentences and paragraphs to express messages, including this article. Researchers view the World Wide Web as a network, and study its characteristics and dynamics.[15][16]
According to Alon, "The cell can be viewed as an overlay of at least three types of networks, which describes protein-protein, protein-DNA, and protein-metabolite interactions."[17] Alon notes that cellular networks, like many human engineered networks, show 'modularity', 'robustness', and 'motifs':
- Modules are subnetworks with a specific function and which connect with other modules often only at one input node and one output node. Modularity facilitates evolutionary adaptation to changing environments, for to produce an adaptation, evolution need tinker with just a few modules rather than with the whole system. Evolution can sometimes 'exapt' existing modules for new functions that contribute to reproductive fitness. For example, the swim bladder purportedly evolved as an adaptation for control of buoyancy later exapted as a respiratory organ in various groups of fish.[18]
- Robustness describes how a network is able to maintain its functionality despite environmental perturbations that affect the components. Robustness also reduces the range of network types that researchers must consider, because only certain types of networks are robust.
- Network motifs offer economy of network design, as the same circuit can have many different uses in cellular regulation, as in the case of autoregulatory circuits and feedforward loops. Nature selects motifs in part for their ability to make networks robust, so systems use motifs that work well over and over again in many different networks.[19] In several well-studied biological networks, the abundance of network motifs — small subnetworks — correlates with the degree of robustness.[20] Networks, like those in cells and those in neural networks in the brain,[21] use motifs as basic building blocks, like multicellular organisms use cells as basic building blocks. Motifs offer biologists a level of simplicity of biological functionality for their efforts to model the dynamics of organized hierarchies of networks.[19]
The view of the cell as an overlay of mathematically-definable (in principle) dynamic networks can reveal how a living system can exist as an improbable, intricate, self-orchestrated dance of molecules.[22] The 'overlay of networks' view also suggests how the concept of self-organized networks can extend to all higher levels of living systems.
Biological system behaviors typically perform one or more evolution-informed functions (e.g., growth), so unraveling the evolutionary history of the networks in a biological system contributes importantly in fully explaining it. Evolutionary history requires knowledge of the mechanisms of both generation and selection.
Examples of biological systems
Examples of biological systems (subsystems) include:
- the biosphere
- ecosystems (e.g., a forest)
- demes (e.g., a local population of a species)
- organisms (e.g., humans; bacteria)
- organs (e.g., brain; the vascular endothelium)
- cells (e.g., epithelial cell)
- metabolic pathways (e.g., glycolysis)
- cellular organelles (e.g., mitochondria)
- genomes (e.g., the entire complement of DNA in an organism, as the ’mouse genome')
- gene complexes (e.g., co-expressing genes)
- genes (e.g., protein blueprints)
History of systems biology
Knowing the historical path(s) that led to a modern scientific program can help in understanding the nature of the program, in particular, its goals and its methodologies, especially through considering the different focuses and approaches that different historians have taken when writing about the same topic.
Systems biology as part of General Systems Theory/Systems Science
Cite error: Invalid <ref> tag; invalid names, e.g. too many
The evolution of molecular biology into systems biology
Westerhoff and Palsson[12] introduce the school of thought “…that systems biology of the living cell has its origin in the expansion of molecular biology to genome-wide analyses.” They point out, however, that molecular biology has an earlier history of systems thinking, in particular in the elucidation of the many molecular regulatory circuits and of their contribution to the logic of the cell. They see technological advances that have permitted rapid collection of large data sets (so-called high-throughput technologies mapping the genome) as upping the scale of that research “…enabling us [molecular biologists] to view the genome as the ‘system’ to study.”
They describe two pathways of enquiry that they see as having merged into modern systems biology of the cell.
One root is the advances in molecular biology that ultimately “…led to efforts toward genome-scale model building to analyze the systems properties of cellular function”: recognition of DNA as the genetic material, identifying the structure of DNA, recombinant technology, automated determination of DNA base sequences, and high-throughput technologies yielding the sequences of entire genomes.
A second, parallel root is the development of non-equilibrium thermodynamics, a predictive mathematical theory for describing the behavior of systems that transfer energy from one place to another or convert energy from one form to another in processes the move towards an irreversible state of stability characterized by randomness or disorder—for biological systems, death. Because living systems produce order and maintain a state of non-randomness, they must carry out processes that keep them in a condition far from the equilibrium of randomness, and they achieve this by transforming energy (and matter and information) taken in from the environment. The non-closed biological system produces order at the expense of the environment it opens to, while the environment becomes more disordered.
Westerhoff and Palsson describe advances in the development of non-equilibrium thermodynamics as presaging molecular and cellular systems biology through ‘quantitative’ integration of system components and through the discovery of principles connecting molecular mechanisms and system behaviors. Westerhoff and Alberghina[24] ask of systems biology: “Did we know it all along?”
Historical milestones in the development of systems biology
Anthony Trewavas, a scientist conducting research in the molecular and systems biology of plant cells, emphasizes the following aspects of the history of systems biology:[25]
- How Rene Descartes’ formulation of reductionism—the concept that the properties of complex objects can be explained by reducing them into their parts and studying the properties of the parts —led biologists to assume that one could explain a complex biological system's behavior from the behavior of its subsystems;
- How reductionism played hand-in-hand with the mechanistic view of reality, leading biologists to view systems as predetermined machines, like clocks;
- How reaction against the reductionist-mechanistic view led to a holistic view of biological systems consistent with Aristotle’s dictum, commonly expressed as "the whole is greater than the sum of its parts";
- How experiments revealed that the averaged behavior of a population did not apply to individual members of the population, however homogeneous the population appeared, complementing experiments revealing non-machine-like toleration of large variability among kindred organisms, organs and cells with respect to behavior and function—each manifesting a so-called norm of reaction that overlapped among individuals;
- How a system’s organization of subsystems itself exerts a level of control and constraint over the range of behaviors available to the subsystems in isolation;
- How a system’s subsystems are organised in hierarchies, where the properties of a subsystem emerge from the dynamic interactions of subsystems at lower levels of the hierarchy;
- How subsystems at lower levels of hierarchy exhibit more variability and how their behavior exhibits more order within the system than without—the higher level emergent properties orchestrate and constrain the behavior of the subsystems generating that emergence;
- How Karl Ludwig von Bertalanffy (1901-1972) recognized that all systems share “…the common property of being composed of interlinked components, in which case they might share similarities in detailed structure and control design”;
- How Michael Polyani (1891-1976) recognized that adjacent levels of a system’s subsystems constrain each other and that upper level behaviors require the lower level behaviors;
- How psychologist and polymath Donald T. Campbell (1916-1996) coined the term “downward causation” to describe higher system level constraints on lower levels, as in constraints on gene expression by higher level subsystems—a prelude to understanding ‘emergent’ systems as having properties or behaviors that “make a difference”, i.e., have causal properties;
- How Claude Bernard, Walter Cannon, and Norbert Weiner established the importance of negative feedback for maintaining stability within large systems, leading to subsequent demonstration of negative feedback at the molecular level;
- How the identification of feed-forward mechanisms led to advances in understanding the features characterizing the design of systems control mechanisms.
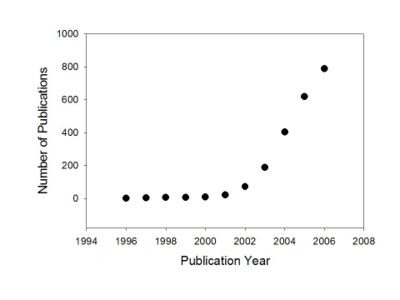
Growth of publications relating systems biology
The pace of research in systems biology greatly accelerated at the beginning of the 21st century.
The figure at left shows the exponential growth of publications relating to or discussing systems biology from 1996 to 2006. The trend for the 1st half of 2007 if continued will put the 2007 figure off the scale of the figure.
The data come from the National Library of Medicine's PubMed database, and probably underestimate annual "systems biology" publication rates by not including articles in journals in many non-biological disciplines. (PubMed searched through the bibliographic database manager, Reference Manager, search term, 'systems biology', searched in 'All Fields.)
Early systems modelers
In 1952, the British neurophysiologists and Nobel laureates Alan Lloyd Hodgkin and Andrew Fielding Huxley constructed a mathematical model of the action potential — the fundamental mechanism underlying communication between nerve cells. In 1960, Denis Noble developed the first computer model of a beating heart.[27]
Computerized mathematical modeling of metabolic networks (Metabolic Control Analysis) began as early as 1960[28]. Metabolic Control Analysis's aim was to quantify the effect of the differing chemical reactions in the network on the concentrations of the network metabolites and on the flow of metabolites through the network, in particular in response to various perturbations (see review by Visser and Heijnen).[29]
Henrik Kacser (1918-1995) is usually credited with publishing the signal paper in Metabolic Control Analysis:
By the end of the 1960s, he [Henrik Kacser] and Jim Burns were working on the theory of how the rates of metabolic pathways were affected by changes in the amounts or activities of pathway enzymes. Their landmark paper, The control of flux (Kacser & Burns, 1973)[30] was presented before a sceptical audience of experimental biochemists in Oxford in 1973. In it they show that the expectation that a metabolic pathway will be controlled by a single pacemaker reaction is a fallacy, and most of the experimental criteria used in the supposed identification of such steps are misleading. Instead, varying amounts of control can be distributed over the enzymes of the pathway, but this is a property of the metabolic system as a whole and cannot be predicted from the characteristics of the enzymes in isolation.[31]
By 1957, Kacser had already revealed himself as a systems biologist, having written: “The problem is therefore the investigation of systems, i.e. components related or organised in a specific way. The properties of a system are in fact more than (or different from) the sum of the properties of its components, a fact often overlooked in zealous attempts to demonstrate additivity of certain phenomonena. It is with these systemic properties that we shall be mainly concerned...”[32]
Systems biology as an evolutionary trend of classical integrative physiology
In the view of physiologists and bioengineers Daniel A. Beard and Marko Vendelin, [33] the term 'systems biology' and 'physiology' synonymize, as they argue:
- "In a nutshell, "systems biology" attempts to realize this [its] promise by measuring and mapping biological interactions within cells, tissues, organs, and organ systems, and predicting how integrated systems made up of many interacting components behave. Readers of the American Journal of Physiology may wish to point out that by such a definition, the term "systems biology" is essentially a synonym for "physiology." While we would not argue with that assessment, we suggest that the increasing attention being paid to the endeavor is at least in part due to the injection of a new term, and associated new techniques and technologies, into the well-established field of integrative physiology. It has been argued that this increase in attention places the discipline of physiology in a position where it will either be "superseded...by systems biology" or will enjoy a renaissance as it leads the way in research in this area..."
Molecular physiologist and biophysicist Kevin Strange."[34] earlier made the comparison:
- ”Physiology and systems biology share the goal of understanding the integrated function of complex, multicomponent biological systems ranging from interacting proteins that carry out specific tasks to whole organisms.”
In writing of systems biology, Strange encourages 21st century physiology departments “to embrace them as essential components” of their field, by ‘them’ referring specifically to “functional genomics, nonmammalian model organisms and computational biology”. He sees a ‘critical mass’-type advantage to a diverse group of physiologists and post-genomics-brand systems biologists working as a group.
Marine biologist, John Bothwell,[35] “…argue[s] that systems biology…retreads the conceptual ground covered by post-Darwinian physiology and resembles nothing more than the application of 19th century physiological reasoning to today’s molecular biological data.” He credits systems biologists for their focus on developing mathematical methodologies for the study of complex cellular networks.
Many others have likened physiology and systems biology. Certainly physiology dates to the beginnings of biology as a discipline, and systems biology dates even to Aristotle. The process of homeostasis, of particular interest to systems biologists, has a long history in physiological studies.[36] In the 19th century, the French physiologist, Claude Bernard, who developed the concept of the ‘internal milieu’, or ‘internal environment’ (e.g., for maintaining blood composition), performed experiments showing animals maintaining their internal milieu within favorable limits—essentially recognizing the process of homeostasis. The early 20th century physiologist Walter Cannon introduced the term 'homeostasis' and demonstrated the homeostatic role of the autonomic nervous system.
The mechanisms of homeostasis still remain central in research physiology, as noted by publications in the American Journal of Physiology journal suite (excluding cell and molecular physiology journals). 43 papers on homeostasis appeared there in 2005 and 2006, the titles of which appear in Appendix A.
Many organ systems physiologists can describe the history, going back to pre-WWII decades, of their fields’ progress in top-down discoveries of organ system components and organization. The mammalian kidney serves as example. In the early decades of the 20th century, the English pharmacologist Arthur Cushny proposed that kidneys produced urine by a dual process of ultrafiltration of blood by the glomeruli and selective reabsorption of the filtrate's constituents by the tubules, and accumulated evidence supported that formulation. Continuing, top-down, to explain the behavior of the organ, renal[37] physiologists developed in the 1930s micropuncture techniques to sample fluid in different segments of the countless ‘nephrons’ — each a unit of kidney anatomy comprising a glomerular filtration apparatus spilling into a variably convoluted tubule, all of which collects in ducts leading to the urinary bladder. That led to discoveries of transport processes that reabsorb and secrete constituents, and to better understanding of how the kidney produced urine of the composition that it did in varying circumstances. They developed quantitative methods for measuring glomerular filtration rate and kidney blood flow, which enabled tracking quantitatively constituents in the filtrate, including in humans. By the time publications had begun to appear by biologists using the term ‘systems biology’, renal physiologists had mathematically modeled numerous kidney systems behaviors, and applied them prevention and treatment of disease in humans.
Physiologists continue to study the systems biology of organs and organ systems, greatly benefiting from bottom-up explorations of molecular and cell biologists, and biochemists and a host of other disciplines.
Perhaps someone will write an integrated article combining the 'History of Physiology' and 'History of Systems Biology'. Constraints on prolixity discourage further development of issues pertaining to the relation of physiology and systems biology in this article.
Modeling in systems biology
For systems biologists, modeling serves as the key to unlocking the system of interest, whether a cell, a species, or an ecosystem.
Reminding us of the story of the six blind men investigating a different part of an elephant and coming up with six different descriptions of the animal, James Haefner[38] wrote:
- One failure of the blind men was to ignore the relations between objects. A seventh man, one sensitive to the importance of testing alternative models, might have said: “Hmmm, ‘tree’, ‘snake’, ‘fan’, ‘spear’, ‘wall’, ‘rope’: It’s a single, big thing with columnar supports and appendages at the ends.” The blind men, especially, need a systems approach, and with respect to the scientific unknown, we are all blind.
To understand a key systems approach, one must know something about model building, or simply, modeling. Models represent (re-present) reality in an abstract form, such as a narrative description (e.g., a theory, conceptualization, rule), a diagrammatic representation (e.g., a graph or network diagram), a physical representation (e.g., a scale model), a 'formal' representation (a mathematical or quantitative description), and simulation models. Most simulation models entail numerical approximations of sets of mathematical expressions potentially describing the system, and ‘stepping-through-time’ with imposed value changes in the states of the system — typically through computer-based computational processing.[39]
Systems biologists use numerous different diagrammatic and mathematical models as keys to understanding biological systems — their logic, say — as keys to explaining, predicting and controlling their behavior.
Mathematical models of a biological system employs sets of equations in describing the system. They posit a series of mathematical expressions (equations) to describe the activity of the system, they comprise analysis of the system by mathematical relations without resort to numerical approximation, and may have dynamic properties by describing the activity of the system over time. They can serve these main functions:
- They allow management of data sets of structures and interactions too large and complex for the human mind to manage without the “exo-cortex” of mathematical formulations, often computer-based for quantitative outputs;
- They allow an iterative process whereby experimental data about a system inspires the model, and the output of the model reveals the need for further experimental research, the results of the latter inspiring changes in the model, which repeats the cycle as the output of the model approaches the realities of the system;
- By failing to fully account for the targeted behavior of the system:
- they raise the possibility of the existence of unknown subsystems of the system that require further experimental work, and may give direction to that further research;
- they inspire refinements of the model to account for the gaps;
- They provide an satisfying and usable ‘explanation’ of the system—an explanation of its properties, functions and behaviors in quantitative terms of its coordinated, dynamically interacting, hierarchically arranged components;
- They enable ‘control’ of the system—the ability to induce a desired behavior or propensity with the appropriate manipulation;
- They enable ‘prediction’ of the behavior of the system—predicting how it will respond to a given set of circumstances, especially predicting novel behaviors of the system.
Despite the large datasets available for many biological systems, especially cell systems, they still fall short in quantity and quality for realizing the full potential of quantitative modeling in applying advanced methodologies for statistical analyses, testing of hypotheses, and estimating the values of equation constants and independent variables.[40] Wellstead et al.[40] note the “enormous challenges to bio-sensing for systems biology” that lie ahead.
Harvard biomathematician and philosopher of science, Richard Levins, speaks to the value of mathematical modeling:
Mathematics is used mostly in modeling in order to predict the outcomes of systems of equations. But it also has another use: educating the intuition so that the obscure becomes obvious. When we abstract from the reality of interest to create mathematical objects, we do this because some questions which would seem intractable now can be grasped immediately. We can look at the fluctuating abundance of insects and conclude ‘‘since these bugs vary by several orders of magnitude during the year and yet remain within bounds from year to year, there must be some density dependence operating (a negative feedback),’’ or see that in a particular patient insulin seems to increase blood sugar and we have to ask, ‘‘where is there a positive feedback loop at work?’’ This kind of qualitative mathematics is essential so that we are not overwhelmed by the sheer numbers of equations and variables of predictive models. The teaching of mathematics to scientists must include the mathematics that aims at understanding rather than solving equations or projecting numbers. [41]
Haefner[38] has provided a classification of mathematical models based on whether the mathematics explicitly represent (a) the mechanisms of the processes of interest, (b) the future states of the system, (c) time as continuous, (d) spatial aspects of the systems, and (e) the effects of random events. Yes/no answers correspond to mathematical structures yielding (a) mechanistic/descriptive models, (b) dynamic/static models, (c)continuous/discrete models, (d) heterogenous/homogenous models of space, and (e) stochastic (random)/deterministic models. A given model might incorporate mathematical structures of any combination of (a) through (e).
Some examples of familiar models
The examples below facilitate learning about modeling heuristically.
A road map of an urban complex

For a given level of detail, a road map represents (re-presents, or models) the structure of the system of roads in a region of land at a given point in time, using the language of graphic illustration. In network parlance, cities and other points of interest (e.g., parks, lakes) serve as nodes and the connections between nodes serve as edges, some of which may indicate one-way connections only. Using the road map, one can control the system of roads in the sense that one can exploit the information to get from one place (node) to another, and predict how long it will take to get there.
The level of detail in a roadmap may not allow determination of the grades of the roads (steepness) or the degrees of curviness or the position of side roads or new roads added since the date of the map.
Systems biologists use maps, for example, to model the structure and interrelationships among biochemical molecules in cellular subsystems, such as the subsystems that convert glucose to usable energy and to other biochemical molecules (e.g., glycogen).
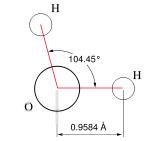
A chemical formula
Anyone reading this far knows the formula, H2O, as a model (representation) for the unit molecule of water. As such it has many uses, in particular in modeling chemical reactions. Other models of water, such as HOH, provide additional information for particular purposes. Note that the model does not give any indication of the 'emergent' properties of interacting collections of H2O molecules, such as liquid water, water ice or steam.
A computer-based flight simulator
For a given level of detail, a computer-based flight simulator represents the multi-subsystem of a flight vehicle system (e.g., airplane), including its control characteristics, and the environment in which the flight vehicle must interact with in order to function stably and exhibit its system behavior of taking off, flying to destination, and landing. The representation (model) uses the languages of engineering, mathematics, computer programs, and dynamical graphics, among others to enable control of the system and to predict its behavior in response to a given set of environmental conditions for comparison with the response to those conditions in real flight—testing the goodness of fit of the model.
Systems biologists use computer-based simulators, for example, to predict the behavior of the human heart in response to drugs.[42]
Newton’s equations
For macroscopic mechanical systems, Newton’s equations represent, or model, the behavior of bodies of mass in motion, allowing predictions of the trajectories of moving masses, including the masses comprising the solar system; allowing calculation of the strength of the mutual attraction of masses and the contribution of each of two masses to that attraction; enabling explanation of macroscopic properties of gases and liquids (e.g., pressure, temperature) from the average effects of microscopic particles interacting in conformity with Newton’s equations; and, enabling engineers to construct vehicles for transporting humans to the moon and back.
(Erwin Schrodinger's wave equation does for subatomic mechanics what Newton's equations did for macroscopic mechanics, and Albert Einstein's General Relativity theory explained away Newton's action at a distance--as models advance in capability.)
Systems biologists use mathematical expressions, often more complex than Newton’s equations, based on empirical data and theoretical principles, in numerous ways to model the behavior of biological systems at all hierarchical levels.[43] See below.
Examples of modeling in systems biology
For examples of web sites with modeling tools for cellular processes, see:[44]
Darwin’s theory of evolution by means of natural and sexual selection
Darwin’s theory, stated conceptually in words, and its 20th century amplification, stated genetically/molecularly often in quantitative terms, represents, or models, the behavior of nature in creating species and varieties sufficiently adapted to their environment to survive long enough to reproduce and care for their offspring and kin. The theory enables understanding of an enormous range of behaviors of animals, plants, unicellular organisms, and cellular and subcellular systems, and has explanatory and predictive value in every biological discipline, and many non-biological disciplines.
Evolutionary principles permeate the discipline of systems biology and have led to the emerging discipline of “evolutionary system biology”[45] A case in point: Determining whether natural selection operating at the molecular level has forged the structure of molecular networks may enhance understanding of their underlying design principles and thereby facilitate design of better predictive models.[46]
Bipedal and Quadrupedal Walking and Running'
In the human organism, walking and running emerge as system behaviors (no subsystem itself walks or runs). The energy cost to the system of those locomotor behaviors defines a property of the system applicable to those behaviors. The energy cost owes to the appropriate forces the system must generate to support itself against gravity and to swing the locomoting limbs to achieve forward motion. Researchers have found that the rate at which the system produces those forces—shorthanded simply to ‘force production’—correlates with the system’s energy cost of locomotion. Thus, if one could develop a mathematical model that predicts force production from readily determined values of variables related to anatomy (e.g., limb length) and motion (e.g., forward speed), that model could then predict the system property of energy cost of locomotion.
Based on the findings of earlier studies, Harvard anthropologist Herman Pontzer[47] developed a mathematical model—viz., an equation—that justified force production as a function of three variables: the rate of muscular force production in the vertical direction, the rate of muscular force production in the horizontal direction, and the rate of muscular force production required to swing the limbs. From empirical data, knowledge of trigonometry and physics (force mechanics) and of muscle physiology, he identified measureable anatomical and motor variables--length and proportion of limbs, speed, frequency of stride, and angle of excursion--that allowed estimation of those required three force variables. Following earlier studies that linked force production with cost of locomotion, he generated the model--the equation--that he hoped would predict the latter from the former. He found that the model well predicted the observed cost of locomotion, somewhat better for running than walking.
Subsequently, Professor Pontzer tested the model in quadrupeds as well as humans.[48] The model proved superior to previous models, extending it to quadrupeds, and confirming the predictive ability of considering the proposed anatomical variables in estimating the rate of force production and energy cost of locomotion.
With the development of quadruped and biped robots for human service, Professor Pontzer's model might help make decisions on energy-cost-effective robot locomotor anatomy and dynamics.
Modeling Noise-Effects on Biochemical Processes and Cell Systems Behavior
Randomness can influence the rate that a gene 'expresses' production of its encoded protein, for example, through random effects on the rate of DNA’s replication as messenger RNA.[49] Biologists refer to that as noise in the gene expression ‘molecular-machine’—fluctuations in expression rate (‘machine output’). A protein repressor of DNA for producing messenger RNA might have a low number of molecules available for the binding to the DNA that effects the repression. In that case, the total time of bound repressor-DNA will vary because of the random nature of molecules diffusing in a solution, which will also affect the time-pattern of messenger RNA production (how often and how long each time). With fewer repressor molecules, random diffusion effects can dictate how rapidly repressor rebinds after diffusing off the DNA, and thereby can influence the total time DNA remains unrepressed and the time-pattern absent repression. Because binding requires two molecules approaching, the spatial distribution of the low numbers of repressor molecules affected by the randomness of its diffusion patterns can produce fluctuating bursts of gene expression---noise.
van Zon and colleagues[49] developed a mathematical model to explain repressor-number-related noise in gene expression based on physical principles and statistical analyses of randomness based on probability theory. They show that their model, with its simplifying assumptions, predicts burst-expression found experimentally, but they admit that the model does not allow for the more complicated effects of random diffusion in cell systems.
van Zon et al.[49] point out that such noise in gene expression as in their model could have significant effects on cellular function, giving specific examples. On that issue, Samilov et al.,[50] in their review of the ‘physiology of noise’, point out that “fluctuations in random biomolecular interactions” [noise] can lead to differences in the behavior of otherwise identical cells (clones) that enable the cells' tissue/organ more choices of adaptive response to environmental changes than a purely noise-free system would have. Noise in biological machinery therefore can have positive effects. Paradigmatic of that, random mutation of gene composition enables evolution by means of natural selection.
Samilov et al.[50] decribe other ways to model noise in biological systems. Rao et al.[51] reference many different modeling approaches to explore the control, rejection, elimination and exploitation of noise, and extensively highlight reasons we need to explain the role of noise in biological systems.
Simulation modeling of an avian influenza pandemic in the United States
To consider the effects of an avian influenza outbreak in the U.S., workers at the Los Alamos National Laboratory, the Fred Hutchinson Cancer Research Center and the School of Public Health and Community Medicine at the University of Washington in Seattle,[52] using a supercomputer and specialized software, simulated the spread of a pandemic influenza viral strain through an artificial U.S. population (281 million people). To perform the simulation, they generated a model with realistic assumptions of transmissibility values, and considered the effect of variable antiviral and vaccine use, and of different modifications of social mobility (e.g., travel restrictions). They describe the basics of their model:
The three basic elements of our national-level simulation model are (i) a previously developed stochastic agent-based model for disease spread at the community level;[53] (ii) detailed U.S. Census demographics and worker flow data for daily commuter traffic at short distances and Bureau of Transportation Statistics data for less frequent long-range travel behavior; and (iii) high-performance parallel computing expertise in modeling millions to billions of particles on hundreds to thousands of processors.
Modeling the mitochondrion
Vo and Palsson[54] reviewed the advances systems biology research has made in explaining the integrated workings of the complex cellular subsystem, the mitochondrion—the cell’s transformer of the external matter and energy taken up by the cell into a usable cellular energy-currency:
- ”With the rapidly increasing number of discovered molecular components [of mitochondria], computational models are also being developed to facilitate the organization and analysis of such data. Computational models of mitochondria have been accomplished with top-down [starting with a theoretical model of the system then tested against the data] and bottom-up [starting with the data and building up to the model] approaches and have been steadily improved in size and scope. Results from top-down methods tend to be more qualitative but are unbiased by prior knowledge about the system. Bottom-up methods often require the incorporation of a large amount of existing data but provide more rigorous and quantitative information, which can be used as hypotheses for subsequent experimental studies.”
To cite one example: Using massive data sets on mitochondrial components and chemical reactions, combined with mathematical assumptions and computer-based mathematical analyse, Vo et al.[55] ‘constructed’ (aka modeled) a network of biochemical pathways that preserved the known interactions among the components. The constructed networked described mitochondrial energy production and other mitochondrial functions closely in accord with experimental results.
The authors recognized the need for more experimental data of mitochondrial components and function to refine the model for predictability.
Modeling by Engineering Synthetic Systems
Systems biologists use advanced and innovative experimental techniques to construct synthetic versions of a subsystem with natural system elements — a unique modeling methodology. They propose a design blueprint for the synthetic subsystem based on empirical data about the system. With appropriate designs or tags, they can then observe and analyze the behavior of the synthetic system, in effect viewing it in isolation from the larger system embedding it. That potentially leads to insights into ways of manipulating the system for desired ends, including inducing novel behaviors of the system and the larger system embedding it.[56]
As articulated by the biologist-physicist team of David Sprinzak and Michael B. Elowitz,[57] synthesizing cellular subsystems has three main goals: (1) to enhance understanding of a natural subsystem by comparing functionality with the synthetic system (e.g., a synthetic metabolic oscillatory circuit) ; (2) to develop non-natural subsystems that potentially provide unique functionality in vivo (e.g., novel gene construction), possibly of value for biotechnology applications; and, (3) to develop synthetic subsystems to replace the natural subsystems in the cell as an exploratory tool to see how it operates in the system-as-a-whole; as the authors write: “. . . perhaps at this stage one can learn more by putting together a simple, if inaccurate, pendulum clock than one can by disassembling the finest Swiss timepiece.”
The brief abstract of Drew Endy’s article on the foundations for a synthetic biology succinctly indicate its potential:[58]
Engineered biological systems have been used to manipulate information, construct materials, process chemicals, produce energy, provide food, and help maintain or enhance human health and our environment. Unfortunately, our ability to quickly and reliably engineer biological systems that behave as expected remains quite limited. Foundational technologies that make routine the engineering of biology are needed. Vibrant, open research communities and strategic leadership are necessary to ensure that the development and application of biological technologies remains overwhelmingly constructive.
The articles cited I this section all describe specific examples of synthetic systems, and the state-of-the-science.
Multi-Level Modeling of the Heart
[...describe work of Denis Noble and colleagues..]
[...Work in progress…More examples to come...]
Emergent properties as ‘materialistic vitalism’
Some cell biologists might find it tempting to see a type of 'vitalism', or 'life force', in living systems, in view of the concept that some high-level features of organisms, including even the activity of living itself, exemplify emergent phenomena — phenomena not explainable after exhaustively studying the properties of each of the chemical processes that occur in the cell in isolation from the organization of the cell system embedding it. [59] One could not explain, for example, the action of an organism fleeing from a predator from a study of the properties of an organism's component subsystems. The properties of the component parts depend on the organization of those parts in the whole system. [4]. Because biologists and their co-scientists can explain emergent properties/phenomena, if only sometimes in principle, by mechanisms that do not transcend interactions of matter and energy, any such ‘vitalism’ properly qualifies only as a ‘materialistic vitalism’.
One example of emergence: When components of signaling pathways, which allow within-cell and between-cell communication, interact to form signaling networks, properties can emerge — such as self-sustaining feedback loops and generation of novel signals — that one cannot explain from the individuated properties of the separate pathways. [60]
For another example, in studying a protein separated from the system it belongs to, one can observe many of its properties, but in so studying the protein one cannot observe any of the properties it has only in the context of the system that embeds it, such as the property of catalyzing a biochemical reaction, or of binding to other proteins to form a functional protein complex. Those properties of the protein emerge in the context of the protein’s environment — how it interacts in the system as a whole. Moreover, those emergent properties may result in effects within the system that, in a feedback way, further alters the properties of the protein in the system, as when a reaction product alters the catalytic properties of the protein.
Every biological system (e.g., a biochemical pathway) resides in a larger, more complex 'parent' system (e.g., an interconnected cycle with feedback) — essentially its environment — that has effects on the 'child' system. Those effects in turn can change the properties and behaviors of the 'child' system's subsystems that would not have existed without the system in its environment — a kind of 'downward causation'. Thus, in an environment of other birds of a feather, a system with flocking behavior may form that one can explain only by studying the behavior of the birds in the flocking-bird organization as a whole.
As Gilbert and Sarkar[4] puts it: “Thus, when we try to explain how the whole system behaves, we have to talk about the context of the whole and cannot get away talking only about the parts.”
Emergent processes have been recognised as, for example, contributing to understanding:
Emergent phenomena appear even in non-biological physical systems. [65] Emergent phenomena attract the attention of cellular neuroscientists; [66] and cognitive scientists [67]. At still higher systems levels, emergent properties appear for example in the behaviour of ant colonies and the concept of swarm intelligence, [68] Artificial systems scientists have simulated emergent phenomena [69] Emergent phenomena in human societies has also received attention. [70]. Biologists even explain the biosphere itself as emergent. [71]
We can elaborate on the principle that knowing the individuated properties of all the components of a system does not enable us to explain the properties of the system-as-a-whole — the opposite principle of the reductionist paradigm prevailing for centuries in physics and chemistry, and even in biology.[72] Firstly, the intrinsic properties of a system’s components do not of themselves allow description of the properties of the whole system, any more than a shoebox of Lego pieces disassembled from its erstwhile airplane reveal the finished product. Rather, the 'organizational dynamics' of the components explains the system, and those dynamics include not only the interrelations among the components themselves, but also interactions among the many different organizational units in the system. [73] Secondly, the living system always operates in some context (its external environment, or surroundings), and that context, in turn, always affects the properties of the system-as-a-whole. For example, nutrient gradients influence the direction a bacterium’s locomotion. The impact of environmental context affects the organization of the components within the system — a 'downward causation'. [74] For another example, environmental signals can activate or suppress a metabolic pathway, reorganizing cellular activity[75] One cannot simply take a living system apart and predict how it will behave.
According to philosopher of science D.M. Walsh: "The constituent parts and processes of a living thing are related to the organism as a whole by a kind of 'reciprocal causation'."[76] In other words, the organization of the components determine the behavior of the system, but that organization arises from more than the set of its internal components. How the whole system behaves as it interacts with its environment determines how those components organize themselves, and so novel properties of the system 'emerge' that characterize neither the environment nor that set of internal components.
For example, the behavior of a human kidney cell depends not only on its cellular physiology, but also on all the properties of the organ (kidney) which constitutes its environment. The kidney's overall structure and function influence the cell’s structure and behavior (e.g., by physical confinement and by cell-to-cell signaling), which in turn influence the organization of its intracellular components. The kidney in turn responds to its environment, namely the organism that it lives in, and that organism responds to its environment, which includes such factors as the availability of particular food items, fresh water, and ambient temperature and humidity.
Systems biologists thus regard emergent properties as arising from from a combination of bottom-up and top-down effects — Walsh's 'reciprocal causation'.
Other basic concepts that systems biologists consider crucial in explaining living systems as-a-whole include 'robustness', 'modularity', and 'networks' — all discussed in sections below. Quantitative modeling and simulation guided by experimental biological data provide the mainstay methodologies of systems biologists.
Emergent systems always display what we recognize as ‘complexity’, a feature we have a difficult time precisely defining. Complex systems appear to require more bits of information (words, sentences, lines of computer code, etc.) to describe than the bits of information in the system itself. [77] The operation of the system itself supplies its own most economical model.
According the paleontologist and origin of life researcher, Robert Hazen, four basic complexity elements underpin emergence in a system: [78]
- a sufficiently large ‘density’ of components, with increasing complexity as the concentration increases, up to a point;
- sufficient interconnectivity of the components, with increasing complexity with greater and more varied types of interconnectivity, up to a point;
- a sufficient energy flow through the system to enable the system’s components to perform the work of interacting in the self-organized way characteristic of the energized system;
- flow of energy through the system in a cyclic manner, presumably facilitating the spatiotemporal patterning characteristic of organized systems.
References
- ↑ Property: quality or trait peculiar to a thing (e.g., mass, volume, ability to reproduce, structure, lifespan, etc.; function: activity specially fitted for a thing (e.g., locomotion, phagocytosis, phototropism, functioning as a molecular motor, energy transduction, etc.); behavior: the activity detected by the observer (e.g., deception, flight, chemotaxis, etc.). The distinctions among those often blur, ‘property’ serving generically in many instances.
- ↑ Kitano H (2002) Systems biology: a brief overview Science 295:1662-1664 PMID 11872829
- ↑ 3.0 3.1 Reid RGB. (2007) Biological Emergences: Evolution by Natural Experiment. A Bradford Book, Cambridge . ISBN 10: 0-262-18257-2
- ↑ 4.0 4.1 4.2 4.3 Gilbert SF, Sarkar S (2000) Embracing complexity: organicism for the 21st century Dev Dyn 219:1-9 PMID 10974666
- Reductionism: “…imagine a materialistic philosophy that claims that all complex entities (including proteins, cells, organisms, ecosystems) can be completely explained by the properties of their component parts. Such an epistemological position is called reductionism, and it is the basis for most of physics and chemistry, and much of biology….. In the reductionist epistemology of science, chemistry and biology are not ultimately independent disciplines, because they will eventually have all their explanations “reduced” to the terms of physics. The reductionist epistemology and methodology is strictly analytical.”
- Holism: ”Let us imagine, though, another (ontologically) materialistic philosophy. Here, complex wholes are inherently greater than the sum of their parts in the sense that the properties of each part are dependent upon the context of the part within the whole in which they operate. Thus, when we try to explain how the whole system behaves, we have to talk about the context of the whole and cannot get away talking only about the parts. This philosophical stance is variously called wholism, holism, or organicism.”
- ↑ Note: Computer simulation: The invention and improvement of high-speed digital computers has changed classical scientific experimentation by providing a medium in which scientists can explore systems by implementing their components, interrelations and interactions in a dynamic reality composed solely of binary digits. The scientist can create a particular reality to implement a physical system from known properties of the system and known principles of the interactions of matter. They can enable the digital reality to ‘imitate’, or ‘simulate’, the system’s behavior in response to simulated changes in environmental conditions, and to output the simulation in various formats readily interpretable by human cognition. Nuclear scientists, aerodynamicists, chemists, economists, as well as biologists, more and more rely on computer simulations to examine the behavior of systems.
- ↑ Weng G, Bhalla US, Iyengar R. (1999) Complexity in biological signaling systems Science 284:92-96 PMID 10102825
- ↑ Keller EF (2005) The century beyond the gene. J Biosci 30:3-10 PMID 15824435
- ↑ Endy D. (2005) Foundations for engineering biology. Nature 438:449-453 PMID16306983
- Review of synthetic biology: “…for biologists, the ability to design and construct synthetic biological systems provides a direct and compelling method for testing our current understanding of natural biological systems.”
- ↑ Westerhoff HV, Kell DB. (2007) The methodologies of systems biology. In: Boogerd FC, Bruggeman FJ, Hofmeyr J-HS, Westerhoff HV (editors) Systems Biology: Philosophical Foundations. Elsevier ISBN 0444520856
- ↑ Bruggeman FJ, Westerhoff HV. (2006) The nature of systems biology. PMID 17113776
- ↑ Burrage K, Hood L, Ragan MA. (2006) Advanced computing for systems biology. Briefings in Bioinformatics 7:390-8.
- See also: TeraGrid project, http://www.teragrid.org
- ↑ 12.0 12.1 Westerhoff HV, Palsson BO (2004) The evolution of molecular biology into systems biology Nature Biotechnology 22:1249-52
- ↑ Bruggeman FJ, Westerhoff HV. (2007) The nature of systems biology. Trends in Microbiology 15:45-50. PMID 17113776
- Outstanding introduction to systems biology — driving forces, challenges, future.
- ↑ Barabasi AL, Oltvai ZN. (2004) Network biology: understanding the cell's functional organization. Nature Reviews Genetics 5:101-113
- ↑ 15.0 15.1 Barabási AL (2002) Linked: The New Science of Networks. Cambridge, Mass: Perseus Pub. ISBN 0-7382-0667-9
- ↑ Watts DJ. (2007) A twenty-first century science. Nature 2007;445:489
- ↑ Alon U (2003) Biological networks: the tinkerer as an engineer. Science 301: 1866-7 PMID 14512615
- ↑ See definition
- ↑ 19.0 19.1 Alon U (2007) Simplicity in biology. Nature 446:497
- ↑ Prill RJ et al. (2004) Dynamic properties of network motifs contribute to biological network organization. PLoS Biol 3: e343
- ↑ Sporns O, Kotter R (2004) Motifs in brain networks. PLoS Biol 2: e369
- ↑ Alon U (2007) An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Chapman and Hall/CRC
- ↑ Skyttner L. (2005) General Systems Theory: Problems, Perspectives, Practice. World Scientific, New Jersey. ISBN 981-256-467-5
- ↑ Westerhoff HV and Alberghina L (2005), 'Systems Biology: Did we know it all along?', in Topics in Current Genetics, Vol. 13: Systems Biology, ed. Alberghina L and Westerhoff HV, pp.3-9. Berlin: Springer-Verlag. ISBN 978-3-540-22968-1
- ↑ Trewavas AJ (2006), A Brief History of Systems Biology: "Every object that biology studies is a system of systems." Francois Jacob (1974). Plant Cell 18:2420-30 PMID 17088606
- ↑ Aristotle On Causality.The Stanford Encyclopedia of Philosophy
- ↑ Denis Noble
- ↑ Chance B, Garfinkel D, Higgins J, Hess B. (1960) Metabolic control mechanisms. 5. A solution for the equations representing interaction between glycolysis and respiration in ascites tumor cells. Journal of Biological Chemistry 235:2426–2439
- ↑ Visser D, Heijnen JJ. (2002) The mathematics of metabolic control analysis revisited. Metabolic Engineering 4:114-123 PMID 12009791
- ↑ Kacser H, Burns JA (1973) The control of flux. Symp. Soc. Exp. Biol. 27, 65-104. Updated: Kacser H, Burns JA, Fell DA. (1995) The control of flux. Biochem. Soc. Trans. 23, 341-366 PMID 7672373
- ↑ Fell DA (1996) Dr Henrik Kacser (1918-1995)
- ↑ Henrik Kacser (1918–1995): an Annotated Bibliography
- ↑ Beard DA, Vendelin M. (2006) Systems biology of the mitochondrion. Am J Physiol Cell Physiol 291:C1101-C1103 PMID 17102034 Full-Text
- ↑ Strange K. The end of "naive reductionism": rise of systems biology or renaissance of physiology? Am J Physiol Cell Physiol 2005;288:C968-C974 PMID 15840560 Full-Text
- ↑ Bothwell JHF (2006) The long past of systems biology. New Phytologist 170:6-10. Link to Full-Text
- ↑ Editorial. (1935) "Homeostasis". The Lancet 225:445-6. Full-Text
- ↑ Note: "renal" means relating to the kidney.
- ↑ 38.0 38.1 Haefner JW. (2005) Modeling Biological Systems: Principles and Applications. Springer, ISBN 0387250115
- ↑ Morowitz HJ (2002, 2004) The Emergence of Everything: How the World Became Complex. Oxford University Press. New York. ISBN 0-19-513513-X Full-Text Available Online at Questia with Subscription
Note from Text: “There are those, myself included, who believe that high-speed computation is to biology and the social sciences what calculus was to physics. Computer science is the mathematically based or formal tool that seems to map best onto the structure of the questions asked by many modern natural sciences, and moves into the domain of the social sciences.” - ↑ 40.0 40.1 Wellstead P, Middleton R, Wolkenhauer O (2006) Feedback Medicine: Control Systems Concepts in Personalised, Predictive Medicine and Combinatorial Intervention. Follow link to full-text
- ↑ Levins R. (2006) Strategies of abstraction. Biol Philos 21:741–755 http://dx.doi.org/10.1007/s10539-006-9052-8 Link to Full-Text]
- ↑ Noble D (2006) “Multilevel Modeling in Systems Biology: From Cells to Whole Organs”. In, Szallasi Z, Stelling J, and Periwal V. (editors) (2006) System Modeling in Cell Biology From Concepts to Nuts and Bolts. A Bradford Book, MIT Press, Cambridge, MA ISBN 0262195488 Chapter 14, pp. 297-312
- ↑ Szallasi Z, Stelling J, and Periwal V. (editors) (2006) System Modeling in Cell Biology From Concepts to Nuts and Bolts. A Bradford Book, MIT Press, Cambridge, MA ISBN 0262195488
- ↑ ST (Signal Transduction) NetWatch: Modeling Tools
- ” Web sites with tools for modeling and simulating cellular processes”
- ↑ Medina M (2005) Genomes, phylogeny, and evolutionary systems biology PNAS 102:6630-5 PMID 15851668
- ↑ Wagner A (2003)Does Selection Mold Molecular Networks? Sci.STKE 2003:e41 PMID 14519859
- ↑ Pontzer H. (2005) A new model predicting locomotor cost from limb length via force production. J Exp Biol 208:1513-24
- ↑ Pontzer H. (2007) Predicting the energy cost of terrestrial locomotion: a test of the LiMb model in humans and quadrupeds. J Exp Biol 210:484-94 PMID 17234618
- ↑ 49.0 49.1 49.2 van Zon JS, Morelli MJ, Tanase-Nicola S, ten Wolde PR. (2006) Diffusion of Transcription Factors Can Drastically Enhance the Noise in Gene Expression. Biophys.J. 91:4350-67 Link to Full-Text
- ↑ 50.0 50.1 Samoilov MS, Price G, Arkin AP. (2006) From Fluctuations to Phenotypes: The Physiology of Noise. Sci.STKE 2006:re17 Link to Full-Text
- ↑ Rao CV, Wolf DM, Arkin AP. (2002) Control, exploitation and tolerance of intracellular noise. Nature 420:231-7 Link to Full-Text
- ↑ Germann TC, Kadau K, Longini IM, Jr., Macken CA. (2006) Mitigation strategies for pandemic influenza in the United States. Proc Natl Acad Sci U S A 103:5935-5940 [PMID 16585506]
- ↑ Note: To give the reader a sense of complexity of the computational technique, we quote the authors:
- To implement this computationally demanding model, we use the high-performance parallel molecular dynamics code “Scalable Parallel Short-range Molecular dynamics” (SPaSM) (8), written in C with message passing interface (MPI) communication. In recent years, this code has been used to model liquid- and solid-phase systems containing millions to billions (9) of atoms, yielding insights into such varied physical processes as dislocation dynamics (10), shock wave-induced plasticity (11), phase transformations (12) in metals, and fluid instabilities (13). The present epidemiological model is readily implemented in SPaSM (and presumably in similar particle-based codes) by replacing the C data structure for atoms (consisting of properties such as particle type, position, velocity, …) with one for persons (age, contact groups, immune system status, …), interatomic force field interactions with a social network and disease transmission model, and atomic classical mechanical trajectories with individual mobility rules (from residence to workplace on a regular basis and occasional long-range travel).
- ↑ Vo TD, Palsson BO. (2007) Building the power house: recent advances in mitochondrial studies through proteomics and systems biology. Am J Physiol Cell Physiol 292:C164-C177 PMID 16885397 Full-Text
- ↑ Vo TD, Greenberg HJ, Palsson BO. Reconstruction and Functional Characterization of the Human Mitochondrial Metabolic Network Based on Proteomic and Biochemical Data. J.Biol.Chem. 2004;279:39532-40 PMID 15205464 Full-Text
- ↑ Hasty J, McMillen D, Collins JJ (2002) Engineered gene circuits. Nature 420:224-30 Link to Full-Text
- ↑ Sprinzak D, Elowitz MB. (2005) 10.1038/nature04335 Reconstruction of genetic circuits. Nature 438:443-448 [PMID 16306982
- ↑ Endy D. (2005) Foundations for engineering biology. Nature 438:449-453 PMID 16306983
- ↑ Berg EL et al (2005) Biological complexity and drug discovery: a practical systems biology approach Syst Biol 152:201-6 PMID 16986261
- ↑ Bhalla US, Iyengar R (1999) Emergent properties of networks of biological signaling pathways., 381-387 PMID 9888852 Link-1 Link-2
- ↑ Tabony J (2006) Microtubules viewed as molecular ant colonies Biol Cell 98:603-17 PMID 16968217
- ↑ e.g. Theise ND, d'Inverno M (2004) Understanding cell lineages as complex adaptive systems Blood Cells Mol Dis 32:17-20 PMID 14757407 and Ruiz i Altaba A, et al (2003) The emergent design of the neural tube: prepattern, SHH morphogen and GLI code Curr Opin Genet Dev 13:513-21 PMID 14550418
- ↑ Jeong H et al(2000) The large scale organisation of metabolic networks Nature 407:651-4 [1]
- ↑ e.g. Grindrod P, Kibble M (2004) Review of uses of network and graph theory concepts within proteomics Expert Rev Proteomics 1:229-38 PMID 15966817 and Ye X, Chu J, Zhuang Y, Zhang S (2005) Multi-scale methodology: a key to deciphering systems biology Front Biosci 10:961-5 PMID 15569634
- ↑ Cho YS et al (2005) Self-organization of bidisperse colloids in water droplets J Am Chem Soc 127:15968-75 PMID 16277541
- ↑ see e.g. Burak Y, Fiete I (2006) Do we understand the emergent dynamics of grid cell activity? J Neurosci 26:9352-4 PMID 16977716
- ↑ e.g. Courtney SM (2004) Attention and cognitive control as emergent properties of information representation in working memory Cogn Affect Behav Neurosci 4:501-16 PMID 15849893
- ↑ Theraulaz G et al (2002) Spatial patterns in ant colonies Proc Natl Acad Sci USA 99:9645-9 PMID 12114538
- ↑ Theraulaz G, Bonabeau E (1999)A brief history of stigmergy Artif Life 5:97-116 PMID 10633572
- ↑ Bonabeau E, Meyer C (2001) Swarm intelligence. A whole new way to think about business Harv Bus Rev 79:106-14 PMID 11345907
- ↑ Field CB, Behrenfeld MJ, Randerson JT, Falkowski P. (1998) Primary Production of the Biosphere: Integrating Terrestrial and Oceanic Components. Science 281:237-40 [http://www.sciencemag.org/cgi/content/full/281/5374/237/ Full-Text
- ↑ Sarkar S. (1998) Genetics and reductionism. Cambridge University Press, Cambridge. ISBN 0521631467
- ↑ Note: For example, physical chemists cannot predict the properties of water from knowledge of its components, hydrogen and oxygen. The way hydrogen and water interact to form H2O, and the way H2O molecules interact, enables the properties of water to 'emerge'.
- ↑ Note: Following up on the example of water, the properties of its environment (e.g., temperature, pressure) affect the way the H2O molecules organize themselves, as ice, or liquid, or steam
- ↑ Note: In relation to downward causation, the environment’s effect can sometimes reach down to the genetic recipe with molecular signals, altering the recipe’s expression and consequently the characteristics of the cells — so-called 'epigenetic' effects. When epigenetic alterations of gene expression occur in the reproductive organs, the system changes can be transmitted to the next generation. See
- Jablonka E, Lamb MJ (2005) Evolution in Four Dimension: Genetic, Epigenetic, Behavioral, and Symbolic Variation in the History of Life. Cambridge: MIT Press
- Gorelick R (2004) Neo-Lamarckian medicine. Med Hypotheses 62:299-303 PMID 14962644
- ↑ Walsh DM (2006) Organisms as natural purposes: the contemporary evolutionary perspective. Stud Hist Philos Biol Biomed Sci 37: 771-91
- ↑ (1991) Zurek WJ (ed) Complexity, Entropy, and the Physics of Information: The Proceedings of the Workshop on the Complexity, Entropy, and the Physics of Information Held May-June, 1989, In Santa Fe, New Mexico. Addison-Wesley Publishing Company, The Advanced Book Program, Redwood City. ISBN 0201515091
- ↑ Hazen RM. (2005) Genesis: The Scientific Quest for Life's Origin. Joseph Henry Press, Washington, DC. ISBN 0309094321
NB: Some links to online full-text sources may require individual subscription to the online source, or access through an institutional subscription. Often the linked page contains at least a summary or abstract of the source if not the full-text.
Recommended reading, notes, and links
Books
- Boogerd FC, Bruggeman FJ, Hofmeyr J-HS, Westerhoff HV (editors) (2007) Systems Biology: Philosophical Foundations. Elsevier ISBN 0444520856
- 360 pages; 14 chapters; Book description and table of contents
- Alon U. (2007) An Introduction to Systems Biology: Design Principles of Biological Circuits. Boca Raton: Chapman and Hall/CRC ISBN 1-58488-642-0
- 301 pages; 12 chapters; 4 appendixes; 23 pages of references; glossary; Table of Contents
- Ashby R. (1947), Principles of the Self-Organizing Dynamic System, Journal of General Psychology, vol. 37, p. 125.
- Banathy B. H. (1992), A Systems View of Education; Concepts and Principles for effective Practice. Eductiona Techology Publications, New Jersey. ISBN 0-87778-245-8
- Bock G and Goode JA (eds). (2002) In Silico" Simulation of Biological Processes, Novartis Foundation Symposium 247. John Wiley & Sons ISBN 0-470-84480-9
- Boulding K., (1985), The World as a Total System, Thousand Oaks, CA, Sage Publications.
- Ellner SP, Guckenheimer J. (2006) Dynamic Models In Biology. Princeton: Princeton University Press. PMID 13-978-0-691-12589-3
- Emery F. E., (ed.), (1969), Systems Thinking: Selected Readings, Penguin, New York
- Flood R., and Carson E., (1988), Dealing with Complexity. An Introduction to the Theory and Application of Systems Science. *New York, Plenum Press
- Kaneko K. (2006) Life: An Introduction to Complex Systems Biology. Berlin: Springer ISBN 3-540-32666-9
- 370 pages; 12 chapters; 127 figures; 16 pages of references; Table of Contents
- Kitano H (editor). (2001) Foundations of Systems Biology. MIT Press ISBN 0-262-11266-3
- Klipp E, Herwig R, Kowald A, Wierling C, Lehrach H. (2005) Systems Biology in Practice. Wiley-VCH: ISBN 3-527-31078-9
- Klir G., (ed.) (1991), Facets of Systems Science, New York, Plenum Press
- Miller J. G., (1978), Living Systems, New York, McGraw Hill Book Company.
- Palsson B. (2006) Systems Biology - Properties of Reconstructed Networks. Cambridge University Press ISBN 9780521859035
- Rapoport A. (1968), General System Theory. In David L. Sills (Ed.). The International Encyclopedia of Social Sciences. Volume 15. New York: Macmillan & The Free Press, 452-458.
- Rosen, R. (1970), Dynamical Systems Theory in Biology. Vol. 1: Stability Theory and its Applications. Wiley, New York.
- Skyttner L. (2005) General Systems Theory: Problems, Perspectives, Practice. New Jersey: World Scientific. ISBN 981-256-467-5
- Szallasi Z, Stelling J, Periwal V (eds). (2006) System Modelling in Cellular Biology: From Concept to Nuts and Bolt. A Bradford Book, The MIT Press ISBN 0-262-19548-8
- 480 pages; 17 Chapters; 36 Contributors; Table of Contents and Full-Texts: Preface; Sample Chapter; Contributors; References
- Schneider ED, Sagan D. (2005) Into the Cool: Energy Flow, Thermodynamics, and Life. Chicago: The University of Chicago Press ISBN 0-226-73937-6
- von Foerster H., Zopf G. W., (eds.), (1962), Principles of Self-Organization, Pergamon, New York
- Weiss, P.A. (1970), Life, Order and Understanding. The Graduate Journal, University of Texas, VolVIII, Supplement.
Articles
- Tyson JJ. 2007 Bringing cartoons to life. Nature 445:823
- Essay: Gives several examples illustrating goal stated in preface: “To understand cells as dynamic systems, mathematical tools are needed to fill the gap between molecular interactions and physiological consequences.”
- Pettinen A, Aho T, Smolander OP, Manninen T, Saarinen A, Taattola KL, Yli-Harja O, Linne ML. (2005) Simulation tools for biochemical networks: evaluation of performance and usability. Bioinformatics 21:357-363
- For examples of computational tools systems biologists use for simulation of living systems: "Simulation of dynamic biochemical systems is receiving considerable attention due to increasing availability of experimental data of complex cellular functions."
- Mandel J, Palfreyman NM, Lopez JA, Dubitzky W. (2004) Representing bioinformatics causality. Briefings in Bioinformatics 5:270-283 PMID 15383213
- ”.. the central concern of bioinformatics becomes the study of dynamical effects in biological systems.” Authors review the advantages and disadvantages of mathematical/computational/graphical/biostatistical tools to study dynamical biological systems: ODE = ordinary differential equations; HFPN = hybrid functional Petrinets; BN = Bayesian networks; DBN = dynamic Bayesian networks; SD = Forrester-type system dynamics
- de Jong H. (2002) Modeling and Simulation of Genetic Regulatory Systems: A Literature Review. Journal of Computational Biology 9:67-103 PMID 11911796
- Indicates tools used to model gene regulatory networks, but applicable to modeling other biological systems. From Abstract: ”This paper reviews formalisms that have been employed in mathematical biology and bioinformatics to describe genetic regulatory systems, in particular directed graphs, Bayesian networks, Boolean networks and their generalizations, ordinary and partial differential equations, qualitative differential equations, stochastic equations, and rule-based formalisms. In addition, the paper discusses how these formalisms have been used in the simulation of the behavior of actual regulatory systems.”
- ”Welcome to the Nature Cell Biology and Nature Reviews Molecular Cell Biology joint Focus on Systems Biology: a User's Guide. This collection of Review-type articles are designed as highly accessible and practical guides to some of the most important approaches and challenges in systems biology.”
Selected external links
- Contains and introduction to, and in-depth discussion of systems biology.
Appendix A
Titles of articles reporting studies on homeostatic mechanisms by physiologists published in 2005 and 2006 in the American Journal of Physiology suite of journals, excluding cellular and molecular physiology:
- Spontaneous, homeostatic, and inflammation-induced sleep in NF-{kappa}B p50 knockout mice
- Attenuation of homeostatic responses to hypotension and glucoprivation after destruction of catecholaminergic rostral ventrolateral medulla neurons
- The Interplay Between Homeostatic Synaptic Plasticity and Functional Dendritic Compartments
- Pituitary adenylyate cyclase activating peptide (PACAP) affects homeostatic sleep regulation in healthy young men
- Temporal Regulation of the Expression Locus of Homeostatic Plasticity
- Gastrointestinal processing of Na+, Cl-, and K+ during digestion: implications for homeostatic balance in freshwater rainbow trout
- Homeostatic functions of vascular endothelial growth factor in adult microvasculature
- Homeostatic responses to caloric restriction: influence of background metabolic rate
- Cardiac endocrine function is an essential component of the homeostatic regulation network: physiological and clinical implications
- Role for the Subthreshold Currents ILeak and IH in the Homeostatic Control of Excitability in Neocortical Somatostatin-Positive Inhibitory Neurons
- Impaired glucose homeostasis and mitochondrial abnormalities in offspring of rats fed a fat-rich diet in pregnancy
- MECHANOTRANSDUCTION AND ENDOTHELIAL CELL HOMEOSTASIS: THE WISDOM OF THE CELL
- Estrogen modulates the mechanical homeostasis of mouse arterial vessels through nitric oxide
- Age-dependent alterations in Ca2+ homeostasis: role of TRPV5 and TRPV6
- Altered contractility and [Ca2+]i homeostasis in phospholemman-deficient murine myocytes: role of Na+/Ca2+ exchange
- Voltage-Gated Channels and Calcium Homeostasis in Mammalian Rod Photoreceptors
- Insulin resistance and whole body energy homeostasis in obese adolescents with fatty liver disease
- "Phosphatonins" and the regulation of phosphorus homeostasis
- Obstructive sleep apnea: wreaking havoc with homeostasis
- Impact of glutamine supplementation on glucose homeostasis during and after exercise
- Role of angiotensin in body fluid homeostasis of mice: effect of losartan on water and NaCl intakes
- Role of glucose in modulating Mg2+ homeostasis in liver cells from starved rats
- Caveolin-1 and regulation of cellular cholesterol homeostasis
- Kidney plays a major role in ammonia homeostasis after portasystemic shunting in patients with cirrhosis
- The kidney in vitamin B12 and folate homeostasis: characterization of receptors for tubular uptake of vitamins and carrier proteins
- Effects of sirolimus on mesangial cell cholesterol homeostasis: a novel mechanism for its action against lipid-mediated injury in renal allografts
- Role of Sgk1 in salt and potassium homeostasis
- Low-carbohydrate diets affect energy balance and fuel homeostasis differentially in lean and obese rats
- Changes in guinea pig gallbladder smooth muscle Ca2+ homeostasis by acute acalculous cholecystitis
- Metabolomic profiling of heat stress: hardening and recovery of homeostasis in Drosophila
- Protective effect of prenatal water restriction on offspring cardiovascular homeostasis in response to hemorrhage
- The vitamin D receptor is not required for fetal mineral homeostasis or for the regulation of placental calcium transfer in mice
- Renal oxidative vulnerability due to changes in mitochondrial-glutathione and energy homeostasis in a rat model of calcium oxalate urolithiasis
- Caffeine-induced arrhythmias in murine hearts parallel changes in cellular Ca2+ homeostasis
- Role of adenosine A1 and A3 receptors in regulation of cardiomyocyte homeostasis after mitochondrial respiratory chain injury
- Targeted inhibition of sarcoplasmic reticulum CaMKII activity results in alterations of Ca2+ homeostasis and cardiac contractility
- Fetal and neonatal exposure to AZT and low-protein diet affects glucose homeostasis: a model with implications for AIDS prevention
- Roles of Slc13a1 and Slc26a1 sulfate transporters of eel kidney in sulfate homeostasis and osmoregulation in freshwater
- Role of high-voltage-activated calcium channels in glucose-regulated {beta}-cell calcium homeostasis and insulin release
- Effects of high-fat diets with different carbohydrate-to-protein ratios on energy homeostasis in rats with impaired brain melanocortin receptor activity
- Reduced PDX-1 expression impairs islet response to insulin resistance and worsens glucose homeostasis
- Hepatic cytochrome P-450 reductase-null mice show reduced transcriptional response to quercetin and reveal physiological homeostasis between jejunum and liver
- Opposing Modifications in Intrinsic Currents and Synaptic Inputs in Post-Traumatic Mossy Cells: Evidence for Single-Cell Homeostasis in a Hyperexcitable Network