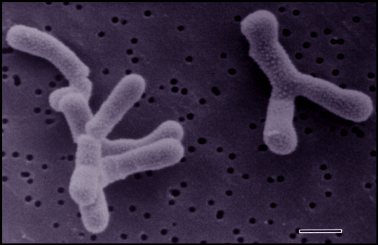
Bifidobacterium bifidum
For the course duration, the article is closed to outside editing. Of course you can always leave comments on the discussion page. The anticipated date of course completion is May 21, 2009. One month after that date at the latest, this notice shall be removed. Besides, many other Citizendium articles welcome your collaboration! |
| Scientific classification | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| ||||||||||||||
| Binomial name | ||||||||||||||
Bifidobacterium bifidum bacteria [1]
Please note this picture will be edited
Description and significance
Bifidobacteria are included in a subsection of bacteria known as probiotics. Probiotics are a natural occurrence in the human body, contributing to the beneficial micro flora in the mouth, intestinal tract, as well as vagina. Bifidobacteria are attributed with a vast array of beneficial physiologic effects which include: digestion promotion, augmentation of the immune system, contribution to the production of lactic and acetic acid, lower serum cholesterol levels, and in some cases are associated with a lower incidence of allergies as well as the promotion of calcium absorption and the synthesis of B-complex vitamins. In addition, bifidobacteria have been utilized as home remedies for diarrhea, vaginitis, yeast infections, as well as irritable bowel syndrome.
Genome structure
Bifidobacterium bifidum has a circular chromosome with a G-C content of approximately 55-67%. The following is the exact 642 base sequence of Bifidobacterium bifidum (ATCC 29521) 16S ribosomal RNA gene as referenced from The National Center for Biotechnology Information (NCBI):
1 gttccacgtg ttccgtgtcg gagctaacgc gttaagcntc ccgcctgggg agtacggccg
61 caaggctaaa actcaaagaa attgacgggg gcccgcacaa gcggcggagc atgcggatta
121 attcgatgca acgcgaagaa ccttacctgg gcttgacatg ttcccgacga cgccagagat
181 ggcgtttcct tcggggcggg ttcacaggtg gtgcatggtc gtcgtcagct cgtgtcgtga
241 gatgttgggt taagtcccgc aacgagcgca accctcgccc cgtgttgcca gcacgttatg
301 gtgggaactc acgggagacc gccggggtta actcggagga aggtggggat gacgtcagat
361 catcatgccc cttacgtcca gggcttcacg catgctacaa tggccggtac agcgggatgc
421 gacatggcga catggagcgg atccctgaaa accggtctca gttcggatcg gagcctgcaa
481 cccggctccg tgaaggcgga gtcgctagta atcgcggatc agcaacgccg cggtgaatgc
541 gttcccgggc cttgtacaca ccgcccgtca agtnatgaaa gtgggcagca cccgaagccg
601 gtggcctaac cccttgtggg atggagccgt ctnaggtgag gc
Cell structure and metabolism
Bifidobacteria are Gram-positive, anaerobic, and nonmotile. The characteristic rod or clubbed shape of Bifidobacterium bifidum varies from 0.5-1.3 μm x 1.5-8 μm. They can be found single or associated in clusters and V-shaped pairs. These bacteria are commonly found curved and in a branched confirmation.
Ecology
Pathology
Application to Biotechnology
Current Research
Effects of galactooligosaccharide and long-chain fructooligosaccharide supplementation during pregnancy on maternal and neonatal microbiota and immunity- a randomized, double-blind, placebo-controlled study.
The effects of prebiotics such as galactooligosaccharides (GOS) and long-chain fructooligosaccharides (lcFOS) that encourage the proliferation of bifidobacteria were examined on maternal gut microbiota during the third trimester of pregnancy and following delivery. This study aimed to determine whether maternal microbiota is passed to newborns via breast-feeding, and, as a secondary outcome, whether maternal gut microbiota affects the fetal immune response. It is known that early infant gut microbiota influence an individual’s susceptibility to developing allergies later in life, although the exact reason and mechanism are unknown. The quantity of bifidobacteria and lactobacilli from the excised population was determined by fluorescent in situ hybridization and quantitative polymerase chain reaction in maternal and infant stool samples. The second objective, to examine the fetal immune response, utilized samples of cord blood by using flow cytometry and cytokine multiplex-array analysis. The results of this study relay that although GOS/lcFOS supplementation during pregnancy has a bifidogenic effect on maternal intestine micro flora, it is not passed on to newborns. Furthermore, there was no correlation between increased maternal bifidobacteria and enhanced fetal immune response.
Exploring the diversity of the bifidobacterial population in the human intestinal tract.
=====While the beneficial aspects of bifidobacteria are widely accepted, the diversity and specific composition of the human intestine micro flora are less understood. Researchers conducted this study to identify specific bifidobacterial populations found in human intestines and fecal samples. Their protocol included plating human intestinal mucosal and fecal samples on selective media and further analyzing molecular data of selected rRNA gene sequences of individual colonies. Their findings clearly indicated that the majority, 704 of the 900 isolated colonies, were bifidobacteria. They further identified the six major species of bifidobacteria isolated from the intestine: B. longum, B. pseudocatenulatum, B. adolescentis, B. pseudolongum, B. breve, and B. bifidum, and two species found primarily in fecal samples, B. dentium and B. animalis subp. lactis. Their research indicated a correlation between age and the microbiota distribution of the intestine. A small selection of species were found exclusively in the adult human gut, while other species were found widely distributed. The study uncovered significant variance between individuals in the composition of fecal samples and intestinal mucosal samples as well as mild variance within the same subject (intrasubject variability) in different regions of the intestine. There were a small number of bifidobacteria that indicated the capacity to broadly colonize, which was based on the number that were able to be isolated from wide ecological distributions.
Isolation of bifidobacteria from breast milk and assessment of the bifidobacterial population by PCR-denaturing gradient gel electrophoresis and quantitative real-time PCR.
References
- ↑ journal citation here