Serratia marcescens
For the course duration, the article is closed to outside editing. Of course you can always leave comments on the discussion page. The anticipated date of course completion is May 21, 2009. One month after that date at the latest, this notice shall be removed. Besides, many other Citizendium articles welcome your collaboration! |
| Serratia marcescens | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
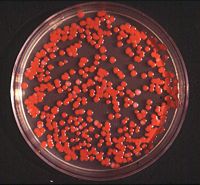 S. marcescens on an agar plate.
| ||||||||||||||
| Scientific classification | ||||||||||||||
| ||||||||||||||
| Binomial name | ||||||||||||||
| Serratia marcescens Bizio 1823 |
Description and significance
Serratia marcescens are gram-negative bacteria which falls under the tribe Klebsielleae and the large family Enterobacteriaceae. It is motile bacteria that are facultative anaerobes.The species, Serratia marcescens is the main pathogen under the genus Serratia. Strains of S marcescens produce prodigiosin, the pigment that gives the bacteria its unique red color. They are rod shaped/ bacillus. It is found in numerous different environments. As a human pathogen, however, it is primarily contracted by hospital patients resulting in urinary and respiratory tract infections. It is also resistant to numerous antibiotics.
Basilio J Anía, MD “Serratia” eMedicine. 1 Oct 2009
Cell Features and Functions
Serratia marcescens are facultative anaerobes, which means they prefer oxygen as a source but can do with out. The red pigment, prodigiosin, is characteristic to the bacteria and is produced by the condensation of an enzyme that forms the pigment. S. marcescens have other distinct features. As opposed to other gram-negative bacteria, they can perform casein hydrolysis; producing metalloproteinases which allow f cell-to-extracellular matrix interaction. For different metabolic process to function S. marcescens also degrade tryptophan and citrate. Citrate is a carbon source for S. marcescens. Researchers have discovered other features of the bacteria, such as adherence and hydrophobicity, and lipopolysaccharide (LPS). In a methyl red test, the bacteria tests negative because it does not perform mixed-acid fermentation. It also produces lactic acid resulting from oxidation and fermentation. Different enzymes have been identified in S. marcescens that contribute to its virulence, such as chitinase, lipase, chloroperoxidase and an extracellular protein, HasA.
Genome structure
The Sanger Institute was funded by the Wellcome Trust, and CNRS, to sequence the genome of Serratia marcescens strain Db11. Dr. Jonathan Ewbank of the Centre d'Immunologie de Marseille Luminy is involved in the sequencing as well. . The genome was completed and consists of a single circular chromosome of 5,113,802 bp with a G+C content of 59.51%. The shotgun reads are still available. The database contains 80,227 reads totalling 51.619 Mb and giving a theoretical coverage of 99.99% of the genome. [1]
Ecology and Habitat
S. marcescens are oomnipresent. They are mesophilic bacteria that prefer moderate or preferably damp environemtns. There optimal temperature is at 38C. Extremely low or high temperature and even pH are lethal to S. marcescens. It can be found in soil, or water, or in an infected host. Ranging from vertebrates, invertebrates, including plants, S. marcescens can infect almost anything. In humans they are mainly found in the urinary and respiratory tract, but any part of the body is susceptible. http://www.ncbi.nlm.nih.gov/sites/entrez?db=genomeprj&cmd=search&term=Serratia%20marcescens