Parainfluenza virus
For the course duration, the article is closed to outside editing. Of course you can always leave comments on the discussion page. The anticipated date of course completion is May 21, 2009. One month after that date at the latest, this notice shall be removed. Besides, many other Citizendium articles welcome your collaboration! |
| Human Parainfluenza Virus | ||
|---|---|---|
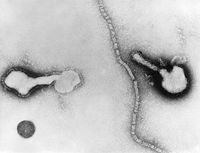 | ||
| Virus classification | ||
|
Description and significance
Upper and lower respiratory tract diseases in humans are mostly caused by Human Parainfluenze Viruses and as a group are called “Parainfluenza.” [1] Human Parainfluenza Viruses (HPIVs) are the second to the Respiratory Synctial Virus (RSV) as a common cause of upper and lower respiratory tract infections especially in infants and young children. [2] It is less common among adults. HPIVs are Paramyxoviruses. Paramyxoviruses belong to Paramyxoviridae family of Mononegavirales order which are negative sense single stranded RNA (-ssRNA) viruses responsible for many diseases in humans and animals. [3] Negative sense RNA first of all makes positive sense RNA through copying their genomes by RNA polymerase. The positive sense RNA molecule is translated into proteins acting as viral mRNA. Thus more –ssRNA are produced when the resultant proteins directs the synthesis of new virions such as capsid proteins and RNA repilcases. [4]
Genome structure
HPIVs are negative sense single stranded RNAs (-ssRNAs).
Cell structure and metabolism
The Human Parainfluenza Viruses are consisting of a nulceocapsid and an envelope with an average diameter of 120-300 nm. The nucleocapsid is a tubelike structure with helical symmetry and 12-18 nm in width. It contains a –ssRNA molecule with 5-8*106 molecular weight, the phoshphopritein P, the major nucleoprotein NP and the RNA polymerase L protein. The L protein is necessary for viral transcription, P proteins takes part in RNA synthesis and NP helps to maintain the genome structure. The envelope is a double layered membrane. The outer layer is covered with spikes which consist of lipoproteins and glycoproteins whereas the inner layer contains non glycosylated matrix protein M. The spikes’s glycoproteins contain hemagglutinin neuraminidase HN and the cell fusion protein F. [5] Viruses acquire the energy from host cells to grow and reproduce. Hemagglutinin in the HPIV’s outer layer spikes attaches the virus to the host cell’s neuraminic acid receptor and F protein facilitates the fusion of virus into the host cell membrane. Since –ssRNA cannot serve as a messenger, L protein, a transcriptase, transcribes the positive sense RNA from the viral genome (-ssRNA). The positive sense RNA serves as mRNA and P protein facilitates the protein synthesis by ribosomes and more –ssRNA strands are produced which are enveloped in double layered membrane. It is stated, “P protein directs the viral protein synthesis and are copied into negative-sense RNA strands which are integrated in the new virions. For envelopment, the virus-specific glycoproteins accumulate in the cell membrane. Assembly is completed by budding of the nucleocapsid through the cell membrane studded with glycoproteins.” [6]
Ecology
HPIVs are pathogens and cause number of diseases since they belong to paramyxoviridae family and all members of this family cause significant diseases. Kelly J. Henrickson says about the paramyxovridae family, “Infact this virus family is one of the most costly in terms of disease burden and economic impact to our planet.” (p : 243) [7] HPIV is strongly affected by environmental conditions such as temperature, pH, humidity and the composition of the medium in which it is stored/grown. As temperature increases from 37°C, its survival chances decreases and it is inactivated within 15 minutes as the temperature approaches to 50°C. It is stable at less temperature such as 4°C and even when it is frozen at -70°C. Freezing might kill the virus or may harm its infectivity but even if very small amount of infectivity is survived, it is enough for the virus to recover. It can be recovered even after 26 years of being frozen. Reagents like bovine serum albumin, skim milk, dimethyl sulfoxide can help the virus in surviving for a long time if they are added into the virus’s culture before freezing. The optimal pH for the Virus’s stability is physiologic pH 7.0 to 8.0 while it loses its stability at ph 3.0 to 3.4, under low humidity and also with virus desiccation. [8]
Pathology
When HPIV infects the body, the respiratory tract secret inflammatory cytokines such as interferon-alpha (IFN), interlukin (IL)-2, (IL-6) and tumor necrosis factor alpha (TNF). Certain chemokines such as RANTES (regulated upon activation, normal T-cell expressed and secreted), macrophage inflammatory protein (MIP)–K have been detected in the nasal secretions. These are responsible for pathological changes in the respiratory tract and mark the onset of illness. The main pathological features of HPIV infection are airway inflammation, necrosis and sloughing of respiratory epithelium, edema, excessive mucus production, and interstitial infiltration of lung. [9]
There are four serotypes of HPIV, HPIV-1, HPIV-2, HPIV-3 and HPIV-4. HPIV-4 has two sub types, HPIV-4a and 4b. Dr. Subhash Chundra Parija discusses the most common diseases caused by different serotypes of HPIVs in correlation with each other: [10]
Croup:. In croup, vocal cords, trachea, bronchi and larynx swell due to the mucous secretions which cause laryngeal obstruction, fever, hoarse barking cough and aspiratory stridor. HPIV-1,2, and 3 are the most frequent causes of croup. They accounting for almost 75% of all cases but only HPIV-1 is responsible for 18% of all croup cases and is considered the most common cause.
Bronchiolitis: HPIV-1 and 3 are the most common cause of bronchiolitis though all four serotypes can be the cause. Bronchiolitis occurs mostly during the first year of life (81% of cases). The major symptoms are expiratory wheezing, rales, trachea retraction, air trapping and fever.
Pneumonias: Like bronchiolitis, HPIV-1 and 3 are the main causes of Pneumonia. Large percentage of hospitalized pneumonia patients is affected by HPIV-3 but each of the two viruses causes almost 10% out outpatient pneumonia cases. Pneumonia’s major symptoms are rales, fever and evidence of pulmonary consolidation.
Tracheobronchitis: More than 25% of the causative agents of tracheobronchitis are HPIVs. All four HPIV types cause the tracheobronchitis but HPIV-3 and 4 are the major causes rather than HPIV-1 and 2.
Other infections: In routine, phyrngitis, conjunctivitis coryza and otitis media can occur with a lower respiratory tract infection or singly by HPIVs. Otitis media mostly causes by HPIV-3.
Infections in immunocompromised patients: The patients who receive intense immunosuppression after they had undergone any organ or bone marrow transplantation had higher risk to develop HPIV infection. Dr. Subhash states, “HPIV-2 causes giant cell pneumonia in persons with severe combined immunodeficiency diseases (SCIDs), and HPIV-3 has been found in persons with SCIDS and acute myeloid leukemia (AML) and in patients who have undergone bone marrow transplantation (BMT). The natural history of HPIV in patients infected with HIV is generally less severe than that in transplant recipients.”
Current Research
A daring treatment and a successful outcome: The need for targeted therapies for pediatric respiratory viruses [11]
This article is about the fact that we need to develop targeted antiviral compounds for HPIVs rather than relying on untested, unproven and partially activated agents. Dr. Stankova treated a child with ribavirin for HPIV-3 infection. She had undergone two stem cell transplantations and was suffering with HPIV-3 infection. Dr. Stankova gave her ribavirin for 10 months. She recovered from the disease and never developed severe respiratory disease again. Since there is no any vaccine or specific medicine for HPIVs treatment, Dr. Stankova wants to take the daring step for using ribavirin as treatment of HPIV-3 infected patients but the authors of the article Patricia DeLaMora and Anne Moscona are against this notion. They argue that ribavirin cannot be used as a treatment for HPIV-3 on regular basis because of its conflicting clinical data and uncertainty in most cases. They say that the child was treated with ribavirin for upper respiratory tract infection with HPIV-3 which also leads to the lower respiratory tract infection but the child did develop lower respiratory tract infection and Dr. Stankova did not explain anything on the role of ribavirin in preventing progression of disease form upper respiratory tract infection to lower respiratory tract infection. They also say, “Published reports do not suggest a strong link between ribavirin use in cases of HPIVs infection and improved out come and there was no difference in mortality for treated and untreated patients suffering with HPIVs.” (p :121, 122)
They suggest that targeted antiviral compounds for HPIVs can be developed. They say that their research shows that binding of viral hemagglutinin-neuraminidase (HN) protein to the host cell is a critical step which activates the fusion (F) protein to fuse the virus into the host cell and if this step is interrupted, host’s cell can be saved from the virus attack. They have identified the specific receptor interacting sites on HPIV-3 and say that now binding inhibitors can be designed specifically to fit into the globular head of HN which will prevent the virus from attaching to the host cell and thus from the attack of HPIV disease. They suggest further that specific peptides can be designed to prevent F protein from activation. They say that, “These several potential therapeutic targets are being actively pursued. It is to be hoped that future dedicated physicians like Stankova et al. will not have to rely on a long shot or a guess, but will have at the ready several strategies to protect and treat children with parainfluenza virus infection.” (p:122)
Human Parainfluenza Type 4 Infections, Canada [12]
In this study Dionne, Moisan and other researchers have discussed some virologic characteristics and clinical menifestations associated with HPIV-4 infections at their hospital in Canada during 2004-2005. They say that this is a rare pathogen and is associated with mild illness but they found the evidences where HPIV-4 caused severe infections. From October 20, 2004 - March 8, 2005, they found 9 respiratory specimens positive for HPIV-4 and it was detected as a severe disease causing virus. Out of these 9 patients, 6 were children with acute bronchiolitis, less than 5/6 months old and one of them, 1.5 month old, had to stay in intensive care unit. Among 3 of the adults, one was 25 years old and other two were above 80. 25 year old young man was suffering with pharyngitis and buccal ulcerative lesions. A 90 year old woman had flulike illness and 84 year old woman was suffering with severe bronchospasm and pulmonary edema. They had to stay in the hospital, were given antimicrobial drugs and were recovered. The reasons for this virus considered to be rare is that it is isolated only occasionally from respiratory tract infections. The previous methods used to detect the virus are not useful due to the virus’s slow growth in LLC-MK2 cell culture and hemadsorption reaction is also occasionally week. It is found lately that RT-PCR method is useful for HPIV-4 detection because due to RT-PCR methods, it is found that HPIV-4 was responsible for 43% of all HPIVs isolated in the laboratory during 2004-2005.
Parainfluenza virus 4 detection in infants. [13]
This study states the importance of the use of real time PCR method for detection of HPIV-4 infections. It says that real time PCR detected the HPIV-4 infections in infants which were undetected by viral cultures. The first child was 5 week old infant with rhinitis and developed tachypnoea, dyspnoea and cyanosis. Her heart beat was 170 per min, her chest X-ray showed hyperinflation with atelectasis of right middle lobe, she had productive cough and rhinorreha with a clear nasal discharge. Other child was 14-month old and was suffering with persistent rhinitis, productive cough, fever, and weight loss due to vomiting. Both children’s nasal wash were tested in cell cultures and real time PCR. Cell culture did not show any viral detection but PCR results were positive for HPIV-4 infection. The one reason for infrequent detection of HPIV-4 is that the cell culture is less sensitive of it than for the other HPIVs and the other reason is that HPIV-4 might be the cause of severe illness like pneumonia and bronchiolitis when its diagnosis is done. This study tells that now real time PCR is using preferentially for the detection of respiratory tract disease because of its rapid and sensitive results. If PCR is not used, almost more than 50% of the respiratory infections may be undiagnosed. It is stated, “PCR is able to expand the number of infections detected and reduce the patients with no viral infection detected. An example of this is human metapneumovirus (hMPV), which is poorly detected by other methods.” (p : 529) It is also discussed In this study that since immune-compromised patients have higher risk to develop life threatening pneumonia due to HPIV-4 infection, it is indispensible to use PCR for quick infection detection so that treatment should be provide on time.
References
- ↑ http://www.nlm.nih.gov/medlineplus/ency/article/001370.htm#Causes,%20incidence,%20and%20risk%20factors
- ↑ http://en.wikipedia.org/wiki/Parainfluenza_virus
- ↑ http://en.wikipedia.org/wiki/Paramyxovirus
- ↑ http://en.wikipedia.org/wiki/Negative-sense_virus
- ↑ http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mmed.section.3128
- ↑ http://www.ncbi.nlm.nih.gov/books/bv.fcgi?rid=mmed.section.3128
- ↑ Kelly J. Henrickson, Parainfluenza Viruses, Clinical Microbiology Reviews, April 2003, Vol 16. No 2, p. 243
- ↑ Kelly J. Henrickson, Parainfluenza Viruses, Clinical Microbiology Reviews, April 2003, Vol 16. No 2, p. 245
- ↑ http://emedicine.medscape.com/article/224708-overview
- ↑ http://emedicine.medscape.com/article/224708-overview Author: Subhash Chandra Parija, MBBS, MD, PhD, FRCPath, Director-Professor of Microbiology, Head of Department of Microbiology, Jawaharlal Institute, Postgraduate Medical Education and Research, India Coauthor(s): Thomas J Marrie, MD, Chair, Professor, Department of Medicine, Division of Infectious Diseases, University of Alberta College of Medicine
- ↑ Patricia DeLaMora and Anne Moscona, A daring treatment and a successful outcome: The need for targeted therapies for pediatric respiratory viruses, Departments of Pediatrics and Microbiology and Immunology, Weill Medical College of Cornell University, New York, USA Pediatr Transplantation 2007: 11: 121–123. http://web.ebscohost.com/ehost/pdf?vid=3&hid=101&sid=7fff0314-3a65-44d6-b372-38f815948b6f%40sessionmgr102
- ↑ Vachon, Dionne, Leblanc, Moisan, Bergeron, and Boivin, Human Parainfluenza Type 4 Infections, Canada, Emerging Infectious Diseases, Vol. 12, No. 11, November 2006. http://web.ebscohost.com/ehost/pdf?vid=7&hid=120&sid=27b19419-b5d9-4a1b-bd1b-9c991dfa6a74%40sessionmgr8
- ↑ Templeton, Bredius, Claas, Kreos, Walther. Parainfluenza virus 4 detection in infants. 19 May 2005. http://web.ebscohost.com/ehost/pdf?vid=5&hid=116&sid=9dfef50b-3d13-4d1c-971b-64a7b9efce04%40sessionmgr102
Be sure to replace "Needs" in "Needs Workgroup" below with a workgroup name. See the "Workgroups" link on the left for a list of workgroups.